3PXG
 
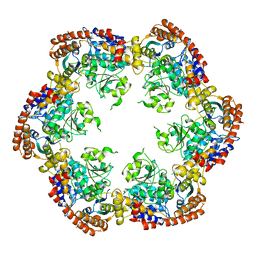 | | Structure of MecA121 and ClpC1-485 complex | | Descriptor: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.654 Å) | | Cite: | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
2PFC
 
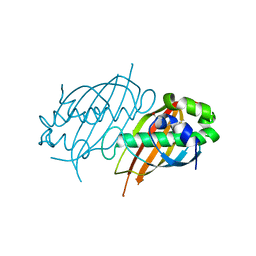 | | Structure of Mycobacterium tuberculosis Rv0098 | | Descriptor: | Hypothetical protein Rv0098/MT0107, PALMITIC ACID | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2007-04-04 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence.
Chem.Biol., 14, 2007
|
|
8D9M
 
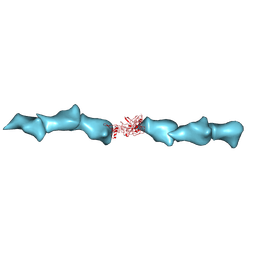 | | Cryo-EM of the OmcZ nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Chan, C.H, Mustafa, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Geobacter OmcZ filaments suggests extracellular cytochrome polymers evolved independently multiple times.
Elife, 11, 2022
|
|
8D4X
 
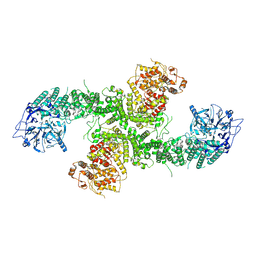 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
3PXI
 
 | | Structure of MecA108:ClpC | | Descriptor: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (6.926 Å) | | Cite: | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
8E0Q
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a C2 symmetric dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
8EWI
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a tetrameric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-10-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
8XPN
 
 | | The Crystal Structure of USP8 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ubiquitin carboxyl-terminal hydrolase 8, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wang, J. | | Deposit date: | 2024-01-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of USP8 from Biortus.
To Be Published
|
|
8WGF
 
 | | The Crystal Structure of JNK3 from Biortus. | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 10, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2023-09-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of JNK3 from Biortus.
To Be Published
|
|
8ZH5
 
 | |
8WGQ
 
 | | The Crystal Structure of L-asparaginase from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-asparaginase | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2023-09-22 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of L-asparaginase from Biortus.
To Be Published
|
|
8YGZ
 
 | | The Crystal Structure of TGF beta R2 kinase domain from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TGF-beta receptor type-2 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of TGF beta R2 kinase domain from Biortus.
To Be Published
|
|
8X70
 
 | | The Crystal Structure of IFI16 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Gamma-interferon-inducible protein 16, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Wang, J. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of IFI16 from Biortus.
To Be Published
|
|
6JWE
 
 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6EF8
 
 | | Cryo-EM of the OmcS nanowires from Geobacter sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C | | Authors: | Wang, F, Gu, Y, Egelman, E.H, Malvankar, N.S. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-10 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers.
Cell, 177, 2019
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
6UQC
 
 | | Mouse IgG2a Bispecific Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, F, Tsai, J.C, Davis, J.H, West, S.M, Strop, P. | | Deposit date: | 2019-10-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design and characterization of mouse IgG1 and IgG2a bispecific antibodies for use in syngeneic models.
Mabs, 12, 2019
|
|
6V7B
 
 | | Cryo-EM reconstruction of Pyrobaculum filamentous virus 2 (PFV2) | | Descriptor: | A-DNA, Structural protein VP1, Structural protein VP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2019-12-08 | | Release date: | 2020-04-01 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a filamentous virus uncovers familial ties within the archaeal virosphere.
Virus Evol, 6, 2020
|
|
5JR3
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Johnson, B.R, Huber, T.D, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone (to be published)
To Be Published
|
|
7ESF
 
 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
5VXX
 
 | | Cryo-EM reconstruction of Neisseria gonorrhoeae Type IV pilus | | Descriptor: | Fimbrial protein, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose | | Authors: | Wang, F, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
5VXY
 
 | | Cryo-EM reconstruction of PAK pilus from Pseudomonas aeruginosa | | Descriptor: | Fimbrial protein | | Authors: | Wang, F, Osinksi, T, Orlova, A, Altindal, T, Craig, L, Egelman, E.H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution.
Structure, 25, 2017
|
|
5WJU
 
 | | Cryo-EM structure of B. subtilis flagellar filaments A39V, N133H | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
7UEG
 
 | |
