4U8Z
 
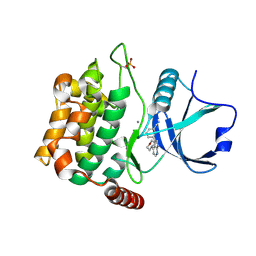 | | Crystal structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06447475) | | Descriptor: | 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, MANGANESE (II) ION, Serine/threonine-protein kinase 24 | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-05 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
9J48
 
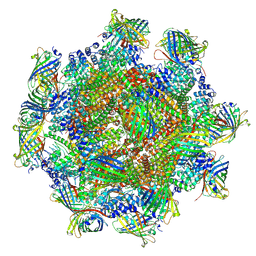 | | GFP bound to 24-mer DARPin-apoferritin model 6c | | Descriptor: | Designed ankyrin repeat proteins,Ferritin heavy chain, N-terminally processed, Green fluorescent protein | | Authors: | Lu, X, Yan, M, Zhang, H.M, Hao, Q. | | Deposit date: | 2024-08-09 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | A large, general and modular DARPin-apoferritin scaffold enables the visualization of small proteins by cryo-EM.
Iucrj, 12, 2025
|
|
8WCJ
 
 | | Crystal structure of GB3 penta mutation L5V/K10H/T16S/K19E/Y33I | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Qin, M.M, Chen, X.X, Zhang, X.Y, Song, X.F, Yao, L.S. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Protein Allostery Study in Cells Using NMR Spectroscopy.
Anal.Chem., 96, 2024
|
|
7FJD
 
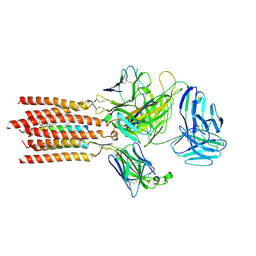 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
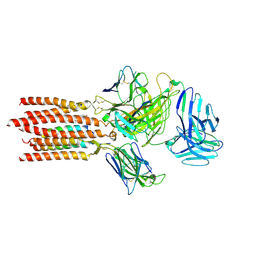 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJF
 
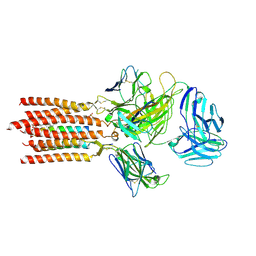 | | Cryo-EM structure of a membrane protein(CS) | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
4W8E
 
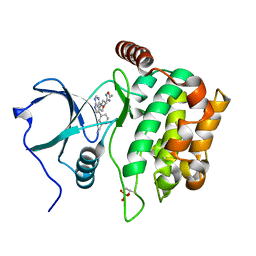 | | Structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06645342) | | Descriptor: | 3-{4-[(2R)-2-(5-methyl-1,2,4-oxadiazol-3-yl)morpholin-4-yl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Serine/threonine-protein kinase 24 36 kDa subunit | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-24 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
5VKC
 
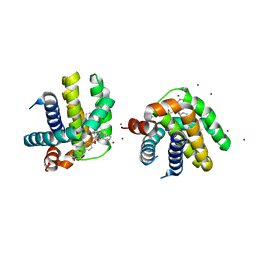 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
4W8D
 
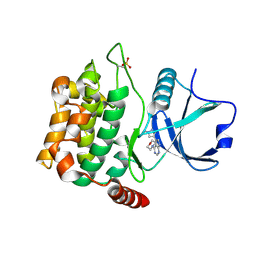 | | Crystal structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06454589). | | Descriptor: | 5-(1-methyl-1H-pyrazol-4-yl)-4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidine, Serine/threonine-protein kinase 24 36 kDa subunit | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-24 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
8HOG
 
 | | Crystal structure of Bcl-2 in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOI
 
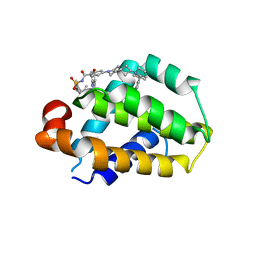 | | Crystal structure of Bcl-2 D103Y in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, FORMIC ACID, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HTS
 
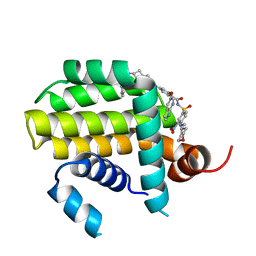 | | Crystal structure of Bcl2 in complex with S-10r | | Descriptor: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
8HTR
 
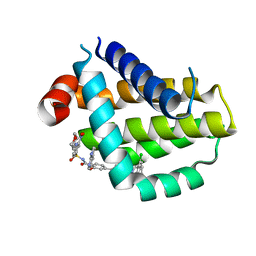 | | Crystal structure of Bcl2 in complex with S-9c | | Descriptor: | 4-[4-[(2~{S})-2-(2-chlorophenyl)pyrrolidin-1-yl]phenyl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
6WPE
 
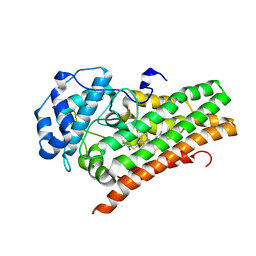 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6X5Y
 
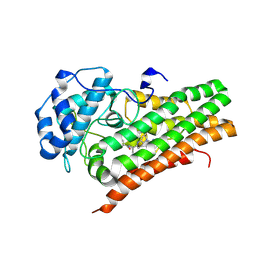 | | IDO1 in complex with compound 4 | | Descriptor: | 4-fluoro-N-{1-[5-(2-methylpyrimidin-4-yl)-5,6,7,8-tetrahydro-1,5-naphthyridin-2-yl]cyclopropyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Utilization of MetID and Structural Data to Guide Placement of Spiro and Fused Cyclopropyl Groups for the Synthesis of Low Dose IDO1 Inhibitors
To Be Published
|
|
6WJY
 
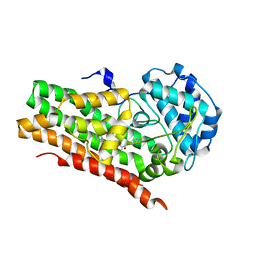 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4-A | | Descriptor: | 3-chloro-N-(3-{(2S)-1-[(4-fluorophenyl)amino]-1-oxopropan-2-yl}bicyclo[1.1.1]pentan-1-yl)benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A, Neumann, L. | | Deposit date: | 2020-04-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of Potent and Orally Available Bicyclo[1.1.1]pentane-Derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6V52
 
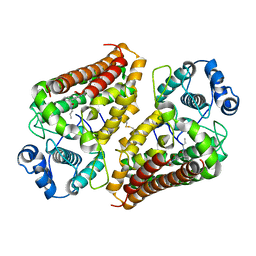 | | IDO1 IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 3-chloro-N-{4-[1-(propylcarbamoyl)cyclobutyl]phenyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Koenig, K.V, Augustin, M.A. | | Deposit date: | 2019-12-03 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strategic Incorporation of Polarity in Heme-Displacing Inhibitors of Indoleamine-2,3-dioxygenase-1 (IDO1).
Acs Med.Chem.Lett., 11, 2020
|
|
8GUG
 
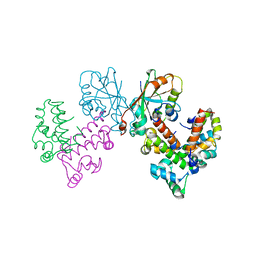 | | Structure of VPA0770 toxin bound to VPA0769 antitoxin in Vibrio parahaemolyticus | | Descriptor: | DUF2384 domain-containing protein, RES domain-containing protein | | Authors: | Song, X.J, Zhang, Y, Xu, Y.Y, Lin, Z. | | Deposit date: | 2022-09-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights of the toxin-antitoxin system VPA0770-VPA0769 in Vibrio parahaemolyticus.
Int.J.Biol.Macromol., 242, 2023
|
|
6VWC
 
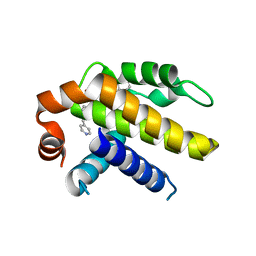 | | Crystal structure of Bcl-xL in complex with tetrahydroisoquinoline-pyridine based inhibitors | | Descriptor: | 6-{8-[(1,3-benzothiazol-2-yl)carbamoyl]-3,4-dihydroisoquinolin-2(1H)-yl}-3-{1-[(pyridin-4-yl)methyl]-1H-pyrazol-4-yl}pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Judge, R.A, Judd, A.S. | | Deposit date: | 2020-02-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Discovery of A-1331852, a First-in-Class, Potent, and Orally-Bioavailable BCL-X L Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
8D84
 
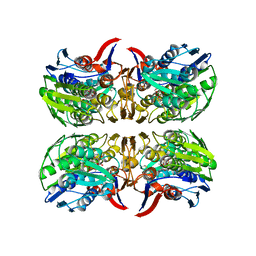 | | E. faecium MurAA in complex with UDP-N-acetylmuramic acid (UNAM) and a covalent adduct of PEP with Cys119 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Enolpyruvate transferase MurAA A149E , identified during adaptation of Enterococcus faecium to daptomycin, increases stability of MurAA-MurG interaction.
J.Biol.Chem., 299, 2023
|
|
3UNP
 
 | | Structure of human SUN2 SUN domain | | Descriptor: | ACETYL GROUP, SUN domain-containing protein 2 | | Authors: | Zhou, Z.C, Greene, M.I. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope
J.Biol.Chem., 287, 2012
|
|
3VEY
 
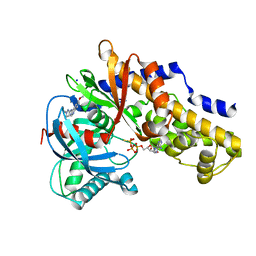 | | glucokinase in complex with glucose and ATPgS | | Descriptor: | 6-methoxy-N-(1-methyl-1H-pyrazol-3-yl)quinazolin-4-amine, Glucokinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3VEV
 
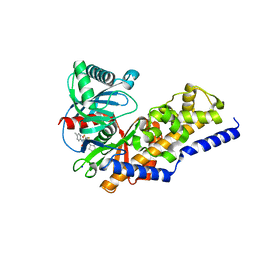 | | Glucokinase in complex with an activator and glucose | | Descriptor: | (2S)-3-cyclopentyl-N-(5-methylpyridin-2-yl)-2-[2-oxo-4-(trifluoromethyl)pyridin-1(2H)-yl]propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3VF6
 
 | | Glucokinase in complex with glucose and activator | | Descriptor: | 6-({(2S)-3-cyclopentyl-2-[4-(trifluoromethyl)-1H-imidazol-1-yl]propanoyl}amino)pyridine-3-carboxylic acid, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
