7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N1M
 
 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Orthorhombic Crystal Form | | Descriptor: | Beta-lactamase OXA-935, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.B, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
7KZ7
 
 | | Crystals Structure of the Mutated Protease Domain of Botulinum Neurotoxin X (X4130B1). | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type X, GLYCEROL, ... | | Authors: | Blum, T.R, Liu, H, Packer, M.S, Xiong, X, Lee, P.G, Zhang, S, Richter, M, Minasov, G, Satchell, K.J.F, Dong, M, Liu, D.R, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phage-assisted evolution of botulinum neurotoxin proteases with reprogrammed specificity.
Science, 371, 2021
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
7KPO
 
 | | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Response regulator, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae.
To Be Published
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L75
 
 | | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Foldase protein PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans.
To Be Published
|
|
7L6L
 
 | | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyribose operon repressor, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12.
To Be Published
|
|
7L6J
 
 | | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative hydrolase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia
To Be Published
|
|
7L6Y
 
 | |
7L6Z
 
 | | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6
To Be Published
|
|
7L71
 
 | |
7L5R
 
 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
7L5V
 
 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Monoclinic Crystal Form | | Descriptor: | Beta-lactamase | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
7L5T
 
 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid
To Be Published
|
|
7KOM
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6 | | Descriptor: | FORMIC ACID, Oxidored_molyb domain-containing protein, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6.
To Be Published
|
|
7KOS
 
 | | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | FORMIC ACID, MALONIC ACID, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7KOU
 
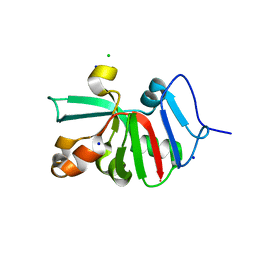 | | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
6U7L
 
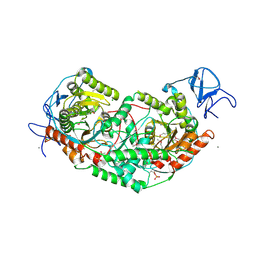 | | 2.75 Angstrom Crystal Structure of Galactarate Dehydratase from Escherichia coli. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactarate dehydratase (L-threo-forming) | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment.
Protein Sci., 29, 2020
|
|
6UOF
 
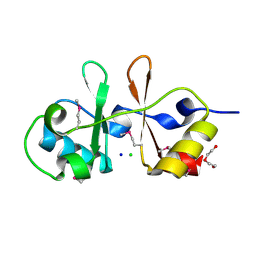 | | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae
To Be Published
|
|
6VBF
 
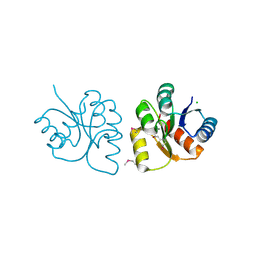 | | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Two-component regulatory system response regulator | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii
To Be Published
|
|
6VJ6
 
 | | 2.55 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus cereus | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, GLYCEROL, Peptidylprolyl isomerase (PrsA) | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus cereus
To Be Published
|
|
6VBB
 
 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
6UE2
 
 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
