3DRR
 
 | | HIV reverse transcriptase Y181C mutant in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3DRS
 
 | | HIV reverse transcriptase K103N mutant in complex with inhibitor R8D | | Descriptor: | 3-chloro-5-[2-chloro-5-(1H-pyrazolo[3,4-b]pyridin-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase/ribonuclease H, p66 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3DRP
 
 | | HIV reverse transcriptase in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
6WVK
 
 | | Cryo-EM structure of Bacillus subtilis RNA Polymerase in complex with HelD | | Descriptor: | DNA helicase IV, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Newing, T, Tolun, G, Oakley, A.J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD.
Nat Commun, 11, 2020
|
|
3CB5
 
 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form A) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CB6
 
 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form B) | | Descriptor: | FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6WVJ
 
 | | Cryo-EM structure of Bacillus subtilis RNA Polymerase elongation complex | | Descriptor: | DNA (5'-D(*TP*GP*TP*CP*GP*GP*GP*CP*GP*TP*CP*CP*GP*CP*GP*CP*GP*CP*C)-3'), DNA (5'-D(P*AP*CP*GP*CP*CP*CP*GP*AP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Newing, T, Tolun, G, Oakley, A.J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD.
Nat Commun, 11, 2020
|
|
3T92
 
 | | Crystal structure of the Taz2:C/EBPepsilon-TAD chimera protein | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ACETONE, HISTONE ACETYLTRANSFERASE P300 TAZ2-CCAAT/ENHANCER-BINDING PROTEIN EPSILON, ... | | Authors: | Bhaumik, P, Maria, M. | | Deposit date: | 2011-08-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into interactions of C/EBP transcriptional activators with the Taz2 domain of p300.
Acta Crystallogr. D Biol. Crystallogr., 70, 2014
|
|
4MON
 
 | | ORTHORHOMBIC MONELLIN | | Descriptor: | MONELLIN | | Authors: | Bujacz, G, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of monellin refined to 2.3 a resolution in the orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
2K8F
 
 | | Structural Basis for the Regulation of p53 Function by p300 | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Bai, Y, Feng, H, Jenkins, L.M, Durell, S.R, Wiodawer, A, Appella, E. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for p300 Taz2-p53 TAD1 Binding and Modulation by Phosphorylation.
Structure, 17, 2009
|
|
1HFK
 
 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with weak sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Lubkowski, J, Palm, G.J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HFJ
 
 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Palm, G.J, Lubkowski, J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3IEC
 
 | |
6BCX
 
 | | mTORC1 structure refined to 3.0 angstroms | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40.
Nature, 552, 2017
|
|
6BCU
 
 | |
3TAM
 
 | |
3V1R
 
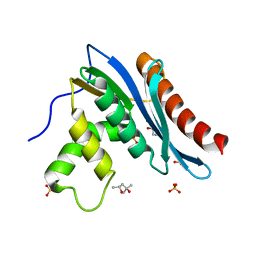 | | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of XMRV with inhibitor beta-thujaplicinol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, ... | | Authors: | Zhou, D, Wlodawer, A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of xenotropic murine leukemia-virus related virus.
J.Struct.Biol., 177, 2012
|
|
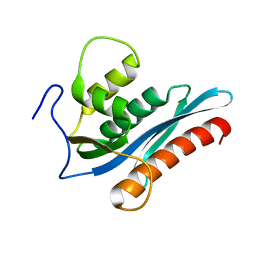
3V1Q
 
 | |
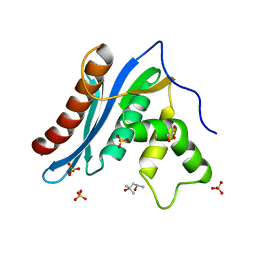
3V1O
 
 | |
1JSR
 
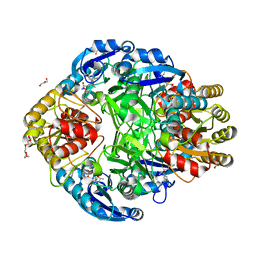 | | CRYSTAL STRUCTURE OF ERWINIA CHRYSANTHEMI L-ASPARAGINASE COMPLEXED WITH 6-HYDROXY-L-NORLEUCINE | | Descriptor: | 6-HYDROXY-L-NORLEUCINE, GLYCEROL, L-asparaginase, ... | | Authors: | Aghaiypour, K, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2001-08-17 | | Release date: | 2002-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Do bacterial L-asparaginases utilize a catalytic triad Thr-Tyr-Glu?
Biochim.Biophys.Acta, 1550, 2001
|
|
1JSL
 
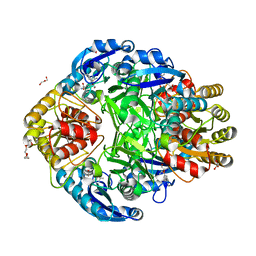 | | Crystal structure of Erwinia chrysanthemi L-asparaginase complexed with 6-HYDROXY-D-NORLEUCINE | | Descriptor: | 6-HYDROXY-D-NORLEUCINE, GLYCEROL, L-asparaginase, ... | | Authors: | Aghaiypour, K, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2001-08-17 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Do bacterial L-asparaginases utilize a catalytic triad Thr-Tyr-Glu?
Biochim.Biophys.Acta, 1550, 2001
|
|
4PGA
 
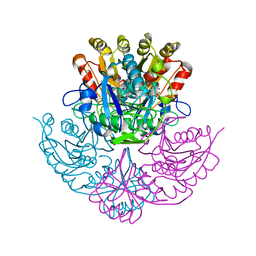 | | GLUTAMINASE-ASPARAGINASE FROM PSEUDOMONAS 7A | | Descriptor: | AMMONIUM ION, GLUTAMINASE-ASPARAGINASE, SULFATE ION | | Authors: | Jakob, C.G, Lewinski, K, Lacount, M.W, Roberts, J, Lebioda, L. | | Deposit date: | 1997-01-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ion binding induces closed conformation in Pseudomonas 7A glutaminase-asparaginase (PGA): crystal structure of the PGA-SO4(2-)-NH4+ complex at 1.7 A resolution.
Biochemistry, 36, 1997
|
|
3I0R
 
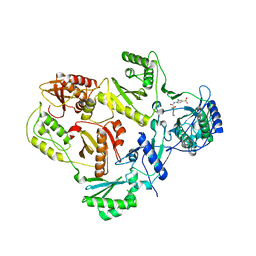 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 3 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6-methyl-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3PGA
 
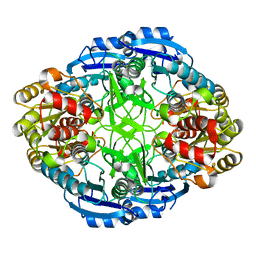 | | STRUCTURAL CHARACTERIZATION OF PSEUDOMONAS 7A GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Ammon, H.L, Copeland, T.D, Swain, A.L. | | Deposit date: | 1994-07-19 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Pseudomonas 7A glutaminase-asparaginase.
Biochemistry, 33, 1994
|
|
3I0S
 
 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 7 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6,8-dichloro-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
