7TRG
 
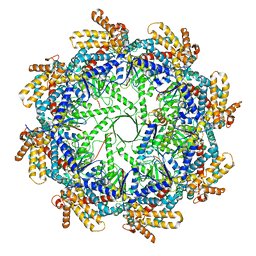 | | The beta-tubulin folding intermediate I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTN
 
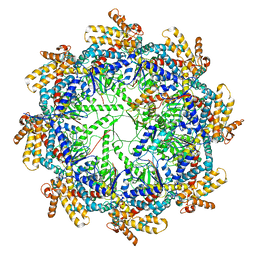 | | The beta-tubulin folding intermediate II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTT
 
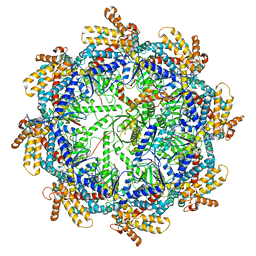 | | The beta-tubulin folding intermediate III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7UG2
 
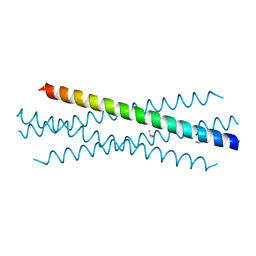 | | Crystal structure of the coiled-coil domain of TRIM75 | | Descriptor: | ACETYL GROUP, ISOPROPYL ALCOHOL, Tripartite motif-containing protein 75 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural studies of the coiled-coil domain of TRIM75 reveal a tetramer architecture facilitating its E3 ligase complex.
Comput Struct Biotechnol J, 20, 2022
|
|
5DH4
 
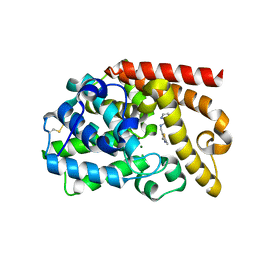 | | PDE10 complexed with 5-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 5-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7W91
 
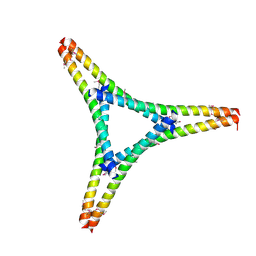 | | Residues 440-490 of centrosomal protein 63 | | Descriptor: | Centrosomal protein of 63 kDa | | Authors: | Yun, H.Y, Ku, B. | | Deposit date: | 2021-12-09 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Architectural basis for cylindrical self-assembly governing Plk4-mediated centriole duplication in human cells.
Commun Biol, 6, 2023
|
|
5DH5
 
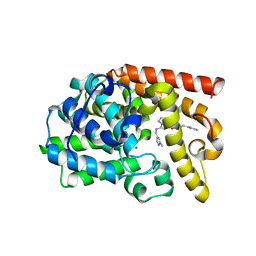 | | PDE10 complexed with N-[(1-methylpyrazol-4-yl)methyl]-5-[[(1S,2S)-2-(2-pyridyl)cyclopropyl]methoxy]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | MAGNESIUM ION, N-[(1-methyl-1H-pyrazol-4-yl)methyl]-5-{[(1S,2S)-2-(pyridin-2-yl)cyclopropyl]methoxy}pyrazolo[1,5-a]pyrimidin-7-amine, ZINC ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8DI4
 
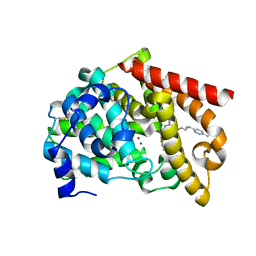 | | Discovery of MK-8189, a highly potent and selective PDE10A inhibitor for the treatment of schizophrenia | | Descriptor: | 2-methyl-6-{[(1S,2S)-2-(5-methylpyridin-2-yl)cyclopropyl]methoxy}-N-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]pyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hayes, R.P, Yan, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Discovery of MK-8189, a Highly Potent and Selective PDE10A Inhibitor for the Treatment of Schizophrenia.
J.Med.Chem., 66, 2023
|
|
6JJJ
 
 | |
7YHH
 
 | |
7YHG
 
 | |
7YHF
 
 | |
7YHI
 
 | |
6KU9
 
 | |
4G0N
 
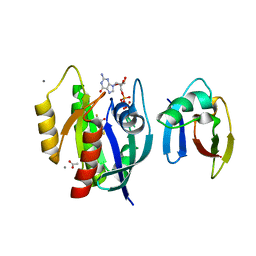 | | Crystal Structure of wt H-Ras-GppNHp bound to the RBD of Raf Kinase | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Fetics, S.K, Kearney, B.M, Buhrman, G, Mattos, C. | | Deposit date: | 2012-07-09 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Allosteric Effects of the Oncogenic RasQ61L Mutant on Raf-RBD.
Structure, 23, 2015
|
|
4G3X
 
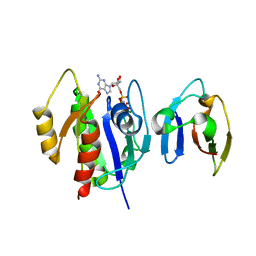 | | Crystal Structure of Q61L H-Ras-GppNHp bound to the RBD of Raf Kinase | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Fetics, S.K, Kearney, B.M, Buhrman, G, Mattos, C. | | Deposit date: | 2012-07-15 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Allosteric Effects of the Oncogenic RasQ61L Mutant on Raf-RBD.
Structure, 23, 2015
|
|
