6YHN
 
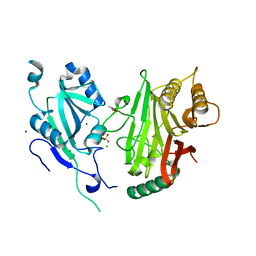 | | Crystal structure of domains 4-5 of CNFy from Yersinia pseudotuberculosis | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, Cytotoxic necrotizing factor, ... | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
6YHM
 
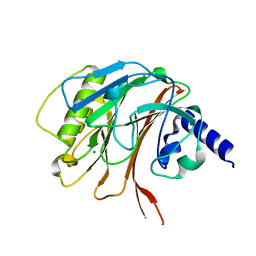 | | Crystal structure of the C-terminal domain of CNFy from Yersinia pseudotuberculosis | | Descriptor: | Cytotoxic necrotizing factor, MAGNESIUM ION | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
6YHK
 
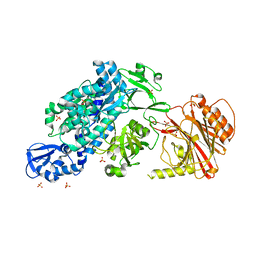 | | Crystal structure of full-length CNFy (C866S) from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, Cytotoxic necrotizing factor, SULFATE ION | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
6ZTH
 
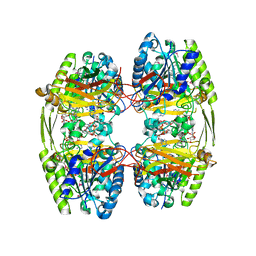 | |
6ZTI
 
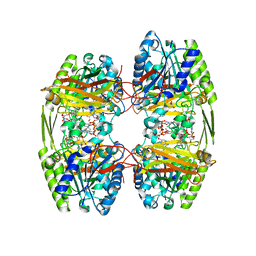 | |
7OZQ
 
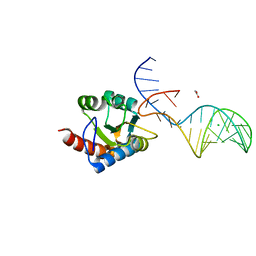 | | Crystal structure of archaeal L7Ae bound to eukaryotic kink-loop | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, CALCIUM ION, ... | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Eukaryotic Box C/D methylation machinery has two non-symmetric protein assembly sites.
Sci Rep, 11, 2021
|
|
7P4U
 
 | |
3JUQ
 
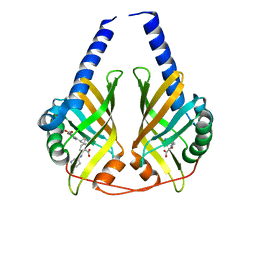 | |
3JUN
 
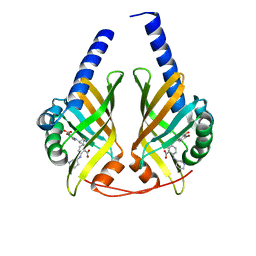 | |
3JUO
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-5-bromo-2-(piperidin-3-ylamino)benzoic acid | | Descriptor: | 5-bromo-2-[(3R)-piperidin-3-ylamino]benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Jain, I.H, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3JUP
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (S)-5-bromo-2-(piperidin-3-ylamino)benzoic acid | | Descriptor: | 5-bromo-2-[(3S)-piperidin-3-ylamino]benzoate, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Jain, I.H, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3JUM
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with 5-bromo-2-((1S,3R)-3-carboxycyclohexylamino)benzoic acid | | Descriptor: | 5-bromo-2-{[(1S,3R)-3-carboxycyclohexyl]amino}benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7QY3
 
 | | Crystal structure of the halohydrin dehalogenase HheG D114C mutant cross-linked with BMOE | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), Putative oxidoreductase, SULFATE ION | | Authors: | Henke, S, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Biocatalytically active and stable cross-linked enzyme crystals of halohydrin dehalogenase HheG by protein engineering
Chemcatchem, 2022
|
|
7QA0
 
 | |
7QA3
 
 | |
2LFF
 
 | | Solution structure of Diiron protein in presence of 8 eq Zn2+, Northeast Structural Genomics consortium target OR21 | | Descriptor: | Diiron protein, ZINC ION | | Authors: | Pires, M, Wu, Y, Mills, J.L, Reig, A, Englander, W, Degrado, W, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Diiron protein in presence of 8 eq Zn2+
To be Published
|
|
7NBW
 
 | |
3DZL
 
 | | Crystal structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-3-oxocyclohexanecarboxylic acid | | Descriptor: | (1R)-3-oxocyclohexanecarboxylic acid, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2008-07-30 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PhzA/B Catalyzes the Formation of the Tricycle in Phenazine Biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
7OC5
 
 | | Alpha-humulene synthase AsR6 from Sarocladium schorii | | Descriptor: | Alpha-humulene synthase AsR6, MAGNESIUM ION, ZINC ION | | Authors: | Schotte, C, Lukat, P, Deuschmann, A, Blankenfeldt, W, Cox, R.J. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Understanding and Engineering the Stereoselectivity of Humulene Synthase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OC6
 
 | | Selenomethionine derivative of alpha-humulene synthase AsR6 from Sarocladium schorii | | Descriptor: | Alpha-humulene synthase asR6, GLYCEROL, SULFATE ION, ... | | Authors: | Schotte, C, Lukat, P, Deuschmann, A, Blankenfeldt, W, Cox, R.J. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-07 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Understanding and Engineering the Stereoselectivity of Humulene Synthase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OC4
 
 | | Alpha-humulene synthase AsR6 from Sarocladium schorii in complex with thiolodiphosphate and a cyclized reaction product. | | Descriptor: | (1E,4E,8E)-2,6,6,9-Tetramethyl-1,4-8-cycloundecatriene, (R,R)-2,3-BUTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Schotte, C, Lukat, P, Deuschmann, A, Blankenfeldt, W, Cox, R.J. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Understanding and Engineering the Stereoselectivity of Humulene Synthase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7T4J
 
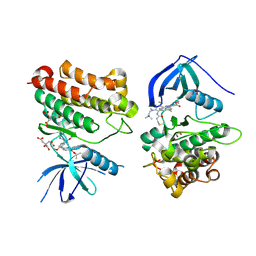 | | Crystal Structure of EGFR_D770_N771insNPG/V948R in complex with TAK-788 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Epidermal growth factor receptor, ... | | Authors: | Skene, R.J, Lane, W, Hu, Y. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7T4I
 
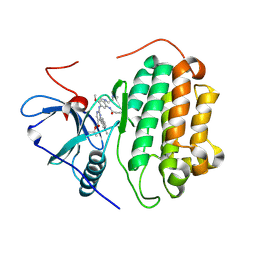 | | Crystal Structure of wild type EGFR in complex with TAK-788 | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Skene, R.J, Lane, W. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
3EX9
 
 | |
6TUM
 
 | |
