8U6Q
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 8-(2-(3-oxo-3-(pyrrolidin-1-yl)propoxy)phenoxy)indolizine-2-carbonitrile (JLJ755), a non-nucleoside inhibitor | | Descriptor: | (4S)-8-{2-[3-oxo-3-(pyrrolidin-1-yl)propoxy]phenoxy}indolizine-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6A
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (JLJ729), a non-nucleoside inhibitor | | Descriptor: | N-(3-{2-[5-chloro-2-(3-chloro-5-cyanophenoxy)phenoxy]ethyl}phenyl)prop-2-enamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6O
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with5-(2-(3-oxo-3-(pyrrolidin-1-yl)propoxy)phenoxy)-2-naphthonitrile (JLJ753), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[3-oxo-3-(pyrrolidin-1-yl)propoxy]phenoxy}naphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6G
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6E
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenethyl)-N-methylacrylamide (JLJ738), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenyl]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6I
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ745), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-[2-(2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U69
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-chloro-5-(4-chloro-2-(2-(5-chloro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)benzonitrile (JLJ334), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(5-chloro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6L
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ748), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-(2-{5-chloro-2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}ethyl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6F
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-(3-chloro-5-cyanophenoxy)phenoxy)ethyl)-N-methylacrylamide (JLJ742), a non-nucleoside inhibitor | | Descriptor: | N-{2-[5-chloro-2-(3-chloro-5-cyanophenoxy)phenoxy]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6T
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ758), a non-nucleoside inhibitor | | Descriptor: | 5-(2-{2-[2-oxo-3-(prop-2-enoyl)-2,3-dihydro-1H-benzimidazol-1-yl]ethoxy}phenoxy)naphthalene-2-carbonitrile, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6N
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)-N,N-dimethylpropanamide (JLJ752), a non-nucleoside inhibitor | | Descriptor: | 3-{2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}-N,N-dimethylpropanamide, Gag-Pol polyprotein, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6K
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ747), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-(2-{2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}ethyl)-N-methylprop-2-enamide, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy)ethyl)-N-methylacrylamide (JLJ736), a non-nucleoside inhibitor | | Descriptor: | N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6S
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 8-(2-(3-morpholino-3-oxopropoxy)phenoxy)indolizine-2-carbonitrile (JLJ757), a non-nucleoside inhibitor | | Descriptor: | (4S)-8-{2-[3-(morpholin-4-yl)-3-oxopropoxy]phenoxy}indolizine-2-carbonitrile, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6C
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 2-chloro-N-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenethyl)acetamide (JLJ732), a non-nucleoside inhibitor | | Descriptor: | 2-chloro-N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenyl]ethyl}acetamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6R
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((2-cyanoindolizin-8-yl)oxy)phenoxy)-N-(2,2-difluoroethyl)propanamide (JLJ756), a non-nucleoside inhibitor | | Descriptor: | 3-(2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)-N-(2,2-difluoroethyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6J
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-((2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ746), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-[2-(5-chloro-2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6M
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((6-chloro-2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ751), a non-nucleoside inhibitor | | Descriptor: | N-[2-(2-{[(4R)-6-chloro-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6H
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8UR9
 
 | | Crystal Structure of the SARS-CoV-2 Main Protease in Complex with Compound 61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proof-of-concept studies with a computationally designed M pro inhibitor as a synergistic combination regimen alternative to Paxlovid.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6OJV
 
 | | Crystal structure of human thymidylate synthase delta(7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-L-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-hydroxy-7H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzoyl}-L-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
6OAV
 
 | | JAK2 JH2 in complex with JAK146 | | Descriptor: | 5-amino-3-[(4-cyanophenyl)amino]-N-phenyl-1H-1,2,4-triazole-1-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Puleo, D.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6OBB
 
 | | JAK2 JH2 in complex with JAK170 | | Descriptor: | 5-amino-N-phenyl-3-[(4-sulfamoylphenyl)amino]-1H-1,2,4-triazole-1-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Puleo, D.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6OE3
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Kudalkar, S.N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and pharmacological evaluation of a novel non-nucleoside reverse transcriptase inhibitor as a promising long acting nanoformulation for treating HIV.
Antiviral Res., 167, 2019
|
|
6OJS
 
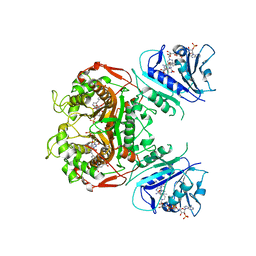 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP, MTX and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Czyzyk, D.J, Anderson, K.S, Jorgensen, W.L, Valhondo, M. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
