8CUK
 
 | |
4H5I
 
 | |
2AYN
 
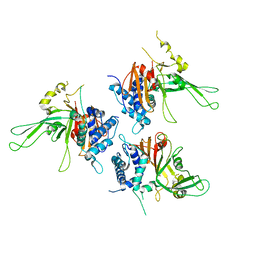 | | Structure of USP14, a proteasome-associated deubiquitinating enzyme | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Hu, M, Li, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-09-07 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
Embo J., 24, 2005
|
|
2AYO
 
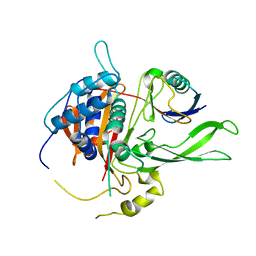 | | Structure of USP14 bound to ubquitin aldehyde | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Hu, M, Li, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-09-07 | | Release date: | 2005-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
Embo J., 24, 2005
|
|
1MYK
 
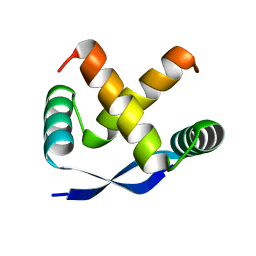 | | CRYSTAL STRUCTURE, FOLDING, AND OPERATOR BINDING OF THE HYPERSTABLE ARC REPRESSOR MUTANT PL8 | | Descriptor: | ARC REPRESSOR | | Authors: | Schildbach, J.F, Milla, M.E, Jeffrey, P.D, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1994-10-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure, folding, and operator binding of the hyperstable Arc repressor mutant PL8.
Biochemistry, 34, 1995
|
|
2F1W
 
 | | Crystal structure of the TRAF-like domain of HAUSP/USP7 | | Descriptor: | CALCIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
4KMO
 
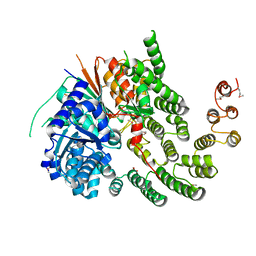 | | Crystal Structure of the Vps33-Vps16 HOPS subcomplex from Chaetomium thermophilum | | Descriptor: | Putative vacuolar protein sorting-associated protein, SULFATE ION, Small conjugating protein ligase-like protein | | Authors: | Baker, R.W, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
Plos One, 8, 2013
|
|
4L9O
 
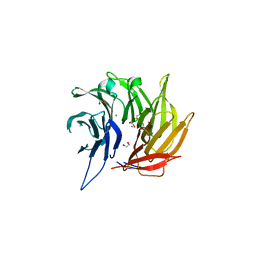 | | Crystal Structure of the Sec13-Sec16 blade-inserted complex from Pichia pastoris | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | McMahon, C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sec16 influences transitional ER sites by regulating rather than organizing COPII.
Mol Biol Cell, 24, 2013
|
|
2F1S
 
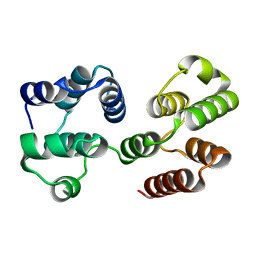 | | Crystal Structure of a Viral FLIP MC159 | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Li, F.-Y, Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Viral FLIP: INSIGHTS INTO FLIP-MEDIATED INHIBITION OF DEATH RECEPTOR SIGNALING.
J.Biol.Chem., 281, 2006
|
|
2F1Z
 
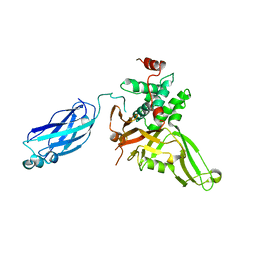 | | Crystal structure of HAUSP | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
2F1Y
 
 | |
2F1X
 
 | |
1JST
 
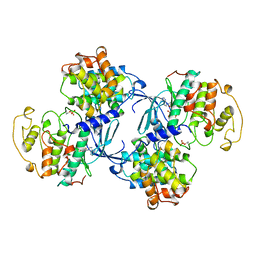 | | PHOSPHORYLATED CYCLIN-DEPENDENT KINASE-2 BOUND TO CYCLIN A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN A, CYCLIN-DEPENDENT KINASE-2, ... | | Authors: | Russo, A.A, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of cyclin-dependent kinase activation by phosphorylation.
Nat.Struct.Biol., 3, 1996
|
|
1BI7
 
 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURE OF THE CDK6-P16INK4A TUMOR SUPPRESSOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, MULTIPLE TUMOR SUPPRESSOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
1BI8
 
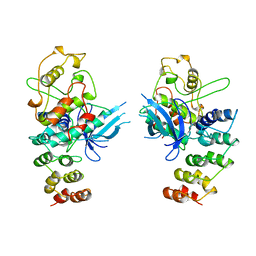 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURES CDK6-P19INK4D INHIBITOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, CYCLIN-DEPENDENT KINASE INHIBITOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
1CEV
 
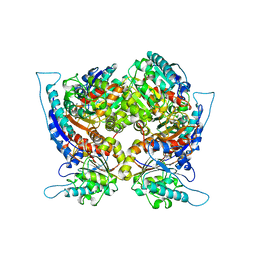 | | ARGINASE FROM BACILLUS CALDOVELOX, NATIVE STRUCTURE AT PH 5.6 | | Descriptor: | MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
3IA3
 
 | |
3KKI
 
 | | PLP-Dependent Acyl-CoA transferase CqsA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CAI-1 autoinducer synthase, MAGNESIUM ION, ... | | Authors: | Kelly, R.C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA.
Nat.Chem.Biol., 5, 2009
|
|
3K8P
 
 | |
3BOC
 
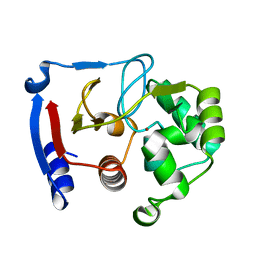 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- zinc bound domain 2 (CDCA1-R2) | | Descriptor: | Cadmium-specific carbonic anhydrase, ZINC ION | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3C5W
 
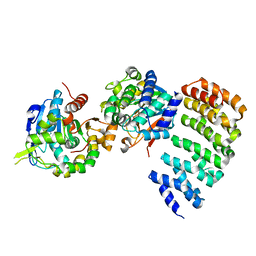 | | Complex between PP2A-specific methylesterase PME-1 and PP2A core enzyme | | Descriptor: | PP2A A subunit, PP2A C subunit, PP2A-specific methylesterase PME-1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
3BOB
 
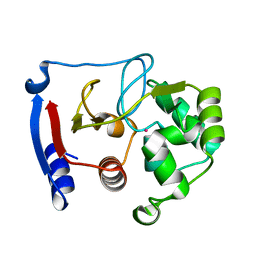 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 | | Descriptor: | CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3HR0
 
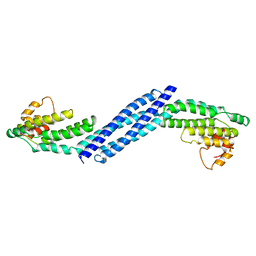 | | Crystal structure of Homo sapiens Conserved Oligomeric Golgi subunit 4 | | Descriptor: | CoG4 | | Authors: | Richardson, B.C, Ungar, D, Nakamura, A, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HQT
 
 | | PLP-Dependent Acyl-CoA Transferase CqsA | | Descriptor: | CAI-1 autoinducer synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kelly, R.C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-06-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA.
Nat.Chem.Biol., 5, 2009
|
|
3BOE
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 with acetate (CDCA1-R2) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
