5H9X
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | Descriptor: | beta-1,3-glucanase | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5H9Y
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | Descriptor: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
4O1V
 
 | | SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, Speckle-type POZ protein | | Authors: | Calabrese, M.F, Watson, E.R, Schulman, B.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer.
Cancer Cell, 25, 2014
|
|
7W33
 
 | | The crystal structure of human CtsL in complex with 14a | | Descriptor: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7W34
 
 | | The crystal structure of human CtsL in complex with 14b | | Descriptor: | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
5T5V
 
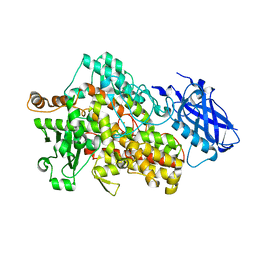 | | LIPOXYGENASE-1 (SOYBEAN) AT 293K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
5Z7G
 
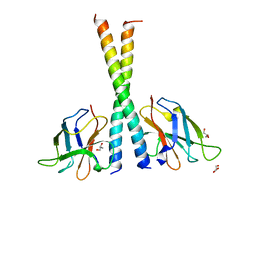 | | Crystal structure of TAX1BP1 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Tax1-binding protein 1 | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q, Wang, Y.L, Hu, S.C. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3H3B
 
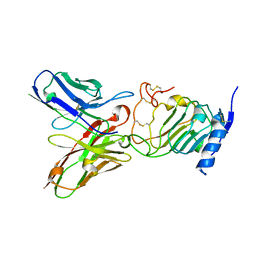 | | Crystal structure of the single-chain Fv (scFv) fragment of an anti-ErbB2 antibody chA21 in complex with residues 1-192 of ErbB2 extracellular domain | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, anti-ErbB2 antibody chA21 | | Authors: | Zhou, H, Liu, Y, Niu, L, Zhu, J, Teng, M. | | Deposit date: | 2009-04-16 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights into the Down-regulation of Overexpressed p185her2/neu Protein of Transformed Cells by the Antibody chA21.
J.Biol.Chem., 286, 2011
|
|
4G3Y
 
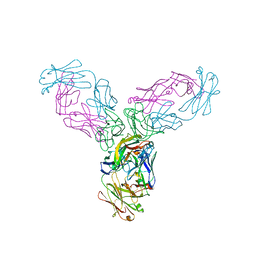 | | Crystal structure of TNF-alpha in complex with Infliximab Fab fragment | | Descriptor: | Tumor necrosis factor, infliximab Fab H, infliximab Fab L | | Authors: | Liang, S.Y, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-07-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab
J.Biol.Chem., 288, 2013
|
|
7CUN
 
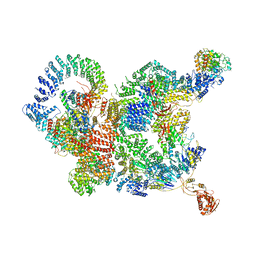 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
5Y3O
 
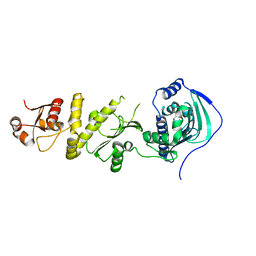 | | Structure of TRAP1 complexed with DN320 | | Descriptor: | 4-chloranyl-1-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Jeong, H, Park, H.K, Kang, S, Kang, B.H, Lee, C. | | Deposit date: | 2017-07-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Paralog Specificity Determines Subcellular Distribution, Action Mechanism, and Anticancer Activity of TRAP1 Inhibitors.
J. Med. Chem., 60, 2017
|
|
6LZE
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11a | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhang, Y, Jing, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6LKO
 
 | |
6MX6
 
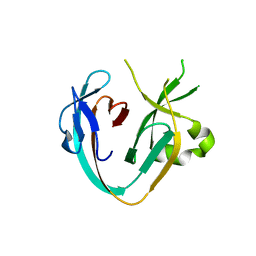 | | The Prp8 intein of Cryptococcus neoformans | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Spliceosomal Prp8 intein at the crossroads of protein and RNA splicing.
Plos Biol., 17, 2019
|
|
7F32
 
 | |
6OWU
 
 | |
6JC3
 
 | | The Cryo-EM structure of nucleoprotein-RNA complex of Newcastle disease virus | | Descriptor: | Nucleocapsid, polyU | | Authors: | Song, X, Shan, H, Zhu, Y, Ding, W, Ouyang, S, Shen, Q.T, Liu, Z.J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Self-capping of nucleoprotein filaments protects the Newcastle disease virus genome.
Elife, 8, 2019
|
|
5GP4
 
 | | Lactobacillus brevis CGMCC 1306 Glutamate decarboxylase | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mei, L, Huang, J. | | Deposit date: | 2016-07-31 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Lactobacillus brevis CGMCC 1306 glutamate decarboxylase: Crystal structure and functional analysis.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
7EXP
 
 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | Descriptor: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
3PWZ
 
 | | Crystal structure of an Ael1 enzyme from Pseudomonas putida | | Descriptor: | Shikimate dehydrogenase 3 | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural and mechanistic analysis of a novel class of shikimate dehydrogenases: evidence for a conserved catalytic mechanism in the shikimate dehydrogenase family.
Biochemistry, 50, 2011
|
|
5EOF
 
 | | Crystal structure of OPTN NTD and TBK1 CTD complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5EP6
 
 | | The crystal structure of NAP1 in complex with TBK1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5EOA
 
 | | Crystal structure of OPTN E50K mutant and TBK1 complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
8Y45
 
 | | Cryo-EM structure of opioid receptor with biased agonist | | Descriptor: | Delta-type opioid receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2024-01-30 | | Release date: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of small-molecule agonist bound delta opioid receptor-G i complex enables discovery of biased compound.
Nat Commun, 15, 2024
|
|
