8VZ1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-409 | | Descriptor: | 4,4'-[(1S,4S,5R)-5-(6-methoxy-3,4-dihydroquinoline-1(2H)-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VYT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-411 | | Descriptor: | 4,4'-[(1R,4R,5S)-5-(2,3-dihydro-1H-indole-1-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VZP
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-403 | | Descriptor: | (1S,2R,4S)-N-(2-hydroxyethyl)-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VZQ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-406 | | Descriptor: | (1S,2R,4S)-N-(2-fluoro-2-methylpropyl)-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NICKEL (II) ION | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VYX
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-410 | | Descriptor: | 4,4'-[(1S,4S,5R)-5-(3,4-dihydroquinoline-1(2H)-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W03
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-1154 | | Descriptor: | (1S,2R,4S)-N-(4-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-13 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W07
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-402 | | Descriptor: | (1R,2S,4R)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.K, Nettles, K.W. | | Deposit date: | 2024-02-13 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5JBM
 
 | | Crystal structgure of Cac1 C-terminus | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Churchill, M.E.A, Liu, W, Zhou, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones.
Elife, 5, 2016
|
|
8C9M
 
 | | HERV-K Gag immature lattice | | Descriptor: | Gag protein | | Authors: | Krebs, A.-S, Liu, H.-F, Zhou, Y, Rey, J.S, Levintov, L, Perilla, J.R, Bartesaghi, A, Zhang, P. | | Deposit date: | 2023-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and conservation of an immature human endogenous retrovirus.
Biorxiv, 2023
|
|
4PQW
 
 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4M7D
 
 | | Crystal structure of Lsm2-8 complex bound to the RNA fragment CGUUU | | Descriptor: | U6 snRNA, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M75
 
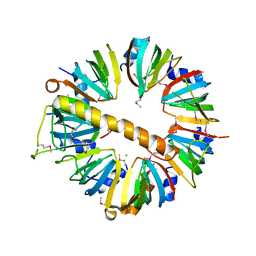 | | Crystal structure of Lsm1-7 complex | | Descriptor: | CHLORIDE ION, U6 snRNA-associated Sm-like protein Lsm1, U6 snRNA-associated Sm-like protein Lsm2, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M77
 
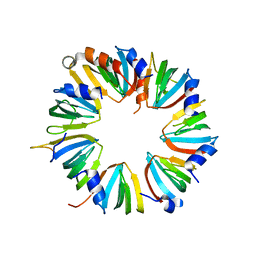 | | Crystal structure of Lsm2-8 complex, space group I212121 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M78
 
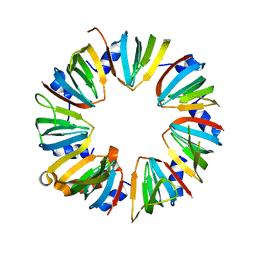 | | Crystal structure of Lsm2-8 complex, space group P21 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
6PDX
 
 | |
6WZT
 
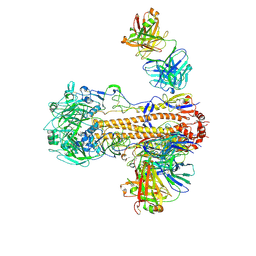 | | CryoEM structure of influenza hemagglutinin A/Victoria/361/2011 in complex with cyno antibody 3B10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cyno antibody heavy chain, ... | | Authors: | Qiu, Y, Zhou, Y, Darricarrere, N. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
6XGC
 
 | |
1YM2
 
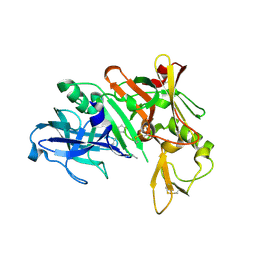 | | Crystal structure of human beta secretase complexed with NVP-AUR200 | | Descriptor: | Beta-secretase 1, NVP-AUR200 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
1YM4
 
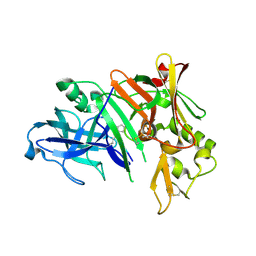 | | Crystal structure of human beta secretase complexed with NVP-AMK640 | | Descriptor: | Beta-secretase 1, NVP-AMK640 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
2DWD
 
 | | crystal structure of KcsA-FAB-TBA complex in Tl+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
2DWE
 
 | | Crystal structure of KcsA-FAB-TBA complex in Rb+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
1KGS
 
 | |
7TAD
 
 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
3A8E
 
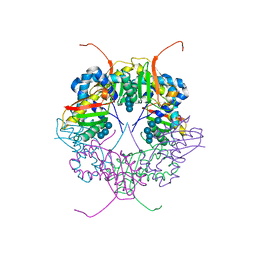 | | The structure of AxCesD octamer complexed with cellopentaose | | Descriptor: | Cellulose synthase operon protein D, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Yao, M, Tanaka, I. | | Deposit date: | 2009-10-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
