8JSO
 
 | | AMPH-bound hTAAR1-Gs protein complex | | Descriptor: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6OHX
 
 | |
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI2
 
 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
4PF9
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-[4-({5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}oxy)phenyl]-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
4PF7
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
4PFC
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-(5-{5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}-2-fluorophenyl)-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
7XKE
 
 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
8SEM
 
 | |
6BA3
 
 | |
3W2D
 
 | | Crystal Structure of Staphylococcal Eenterotoxin B in complex with a novel neutralization monoclonal antibody Fab fragment | | Descriptor: | Enterotoxin type B, Monoclonal Antibody 3E2 Fab figment heavy chain, Monoclonal Antibody 3E2 Fab figment light chain, ... | | Authors: | Liang, S.Y, Hu, S, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the neutralization and specificity of Staphylococcal enterotoxin B against its MHC Class II binding site.
MAbs, 6, 2014
|
|
6M8M
 
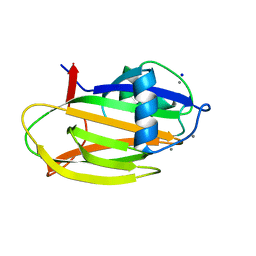 | | PA14 sugar-binding domain from RTX adhesin | | Descriptor: | CALCIUM ION, Putative large adhesion protein (Lap) involved in biofilm formation, SODIUM ION, ... | | Authors: | Vance, T.D.R, Conroy, B, Davies, P.L. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and functional analysis of a bacterial adhesin sugar-binding domain.
Plos One, 14, 2019
|
|
2N8K
 
 | | Chemical Shift Assignments and Structure Determination for spider toxin, U33-theraphotoxin-Cg1c | | Descriptor: | U33-theraphotoxin-Cg1b | | Authors: | Chin, Y.K.-Y, Pineda, S.S, Mobli, M, King, G.F. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2N6N
 
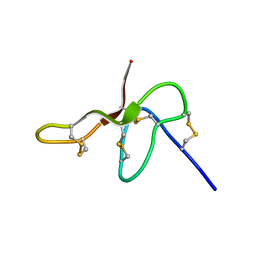 | | Structure Determination for spider toxin, U4-agatoxin-Ao1a | | Descriptor: | U4-agatoxin-Ao1a | | Authors: | Pineda, S.S, Chin, Y.K.-Y, Mobli, M.S, King, G.F. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2N6R
 
 | |
2I0J
 
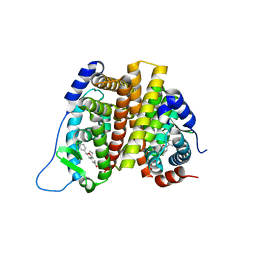 | | Benzopyrans are Selective Estrogen Receptor beta Agonists (SERBAs) with Novel Activity in Models of Benign Prostatic Hyperplasia | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor alpha | | Authors: | Wang, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Benzopyrans are selective estrogen receptor Beta agonists with novel activity in models of benign prostatic hyperplasia.
J.Med.Chem., 49, 2006
|
|
2I0G
 
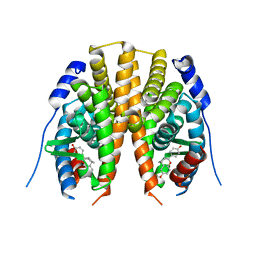 | | Benzopyrans are Selective Estrogen Receptor beta Agonists (SERBAs) with Novel Activity in Models of Benign Prostatic Hyperplasia | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor beta | | Authors: | Wang, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Benzopyrans are selective estrogen receptor Beta agonists with novel activity in models of benign prostatic hyperplasia.
J.Med.Chem., 49, 2006
|
|
8JGK
 
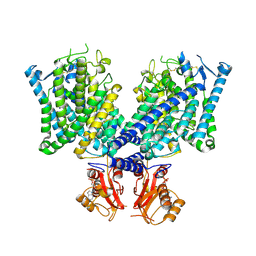 | | Cryo-EM structure of mClC-3 with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JEV
 
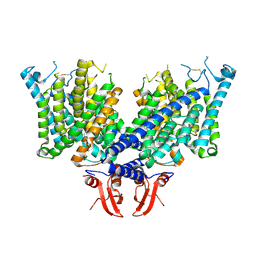 | | Cryo-EM structure of apo state mClC-3 | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGV
 
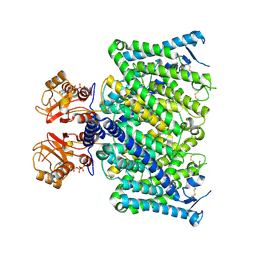 | | Cryo-EM structure of mClC-3_I607T with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGJ
 
 | | Cryo-EM structure of mClC-3 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGS
 
 | | Cryo-EM structure of apo state mClC-3_I607T | | Descriptor: | H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGL
 
 | | Cryo-EM structure of mClC-3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
