8VUQ
 
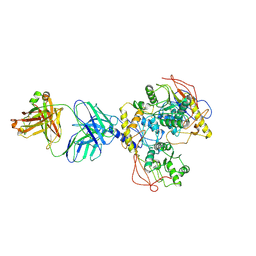 | |
8VUR
 
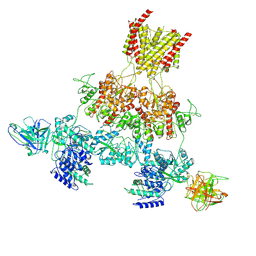 | | Human GluN1-2A with IgG 003-102 WT conformation | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 2024
|
|
8VUU
 
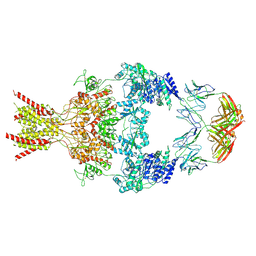 | | Human GluN1-2B with Fab 007-168 | | Descriptor: | 007-168 Heavy, 007-168 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 2024
|
|
8VUV
 
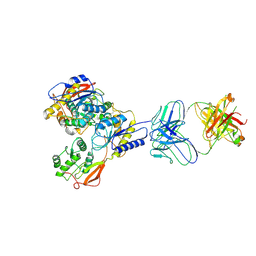 | |
3QEL
 
 | | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2B in complex with ifenprodil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors.
Nature, 475, 2011
|
|
3QEK
 
 | | Crystal structure of amino terminal domain of the NMDA receptor subunit GluN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NMDA glutamate receptor subunit, POTASSIUM ION, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors.
Nature, 475, 2011
|
|
8VUH
 
 | | Human GluN1-2A IgG 003-102 splayed conformation | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 2024
|
|
3QEM
 
 | | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2B in complex with Ro 25-6981 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors.
Nature, 475, 2011
|
|
8USW
 
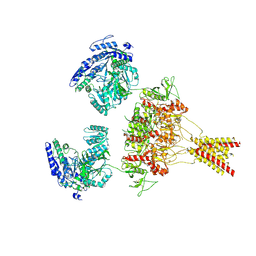 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
8UUE
 
 | |
5FXH
 
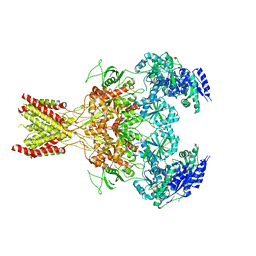 | | GluN1b-GluN2B NMDA receptor in non-active-1 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXI
 
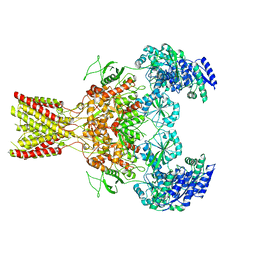 | | GluN1b-GluN2B NMDA receptor structure in non-active-2 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXK
 
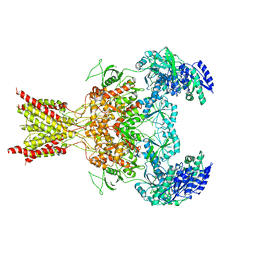 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXG
 
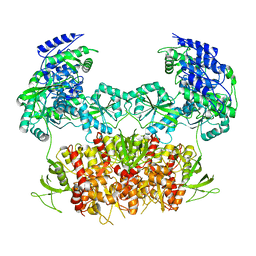 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
8USX
 
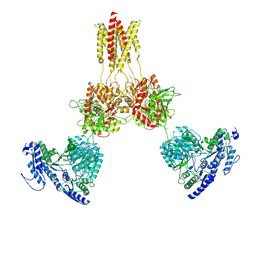 | | Glycine-bound GluN1a-3A NMDA receptor | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 3A | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
9ARI
 
 | |
9ARG
 
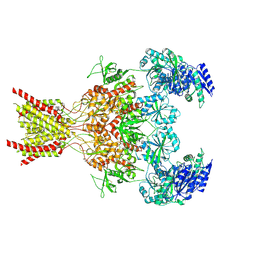 | |
9ARH
 
 | | Rat GluN1-GluN2B NMDA receptor channel in complex with glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Molecular mechanism of ligand gating and opening of NMDA receptor.
Nature, 632, 2024
|
|
6USV
 
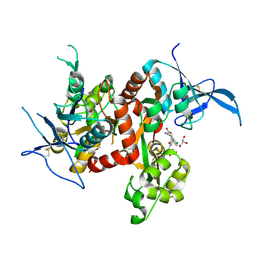 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6VAL
 
 | |
6VAI
 
 | |
6UZ6
 
 | |
6USU
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with L689,560 and glutamate | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6UZG
 
 | |
6UZX
 
 | | Crystal structure of GLUN1/GLUN2A-4M mutant ligand-binding domain in complex with glycine and UBP791 | | Descriptor: | (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid, GLYCEROL, GLYCINE, ... | | Authors: | Wang, J.X, Furukawa, H. | | Deposit date: | 2019-11-15 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors.
Nat Commun, 11, 2020
|
|
