2AJH
 
 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase complexes with methionine | | Descriptor: | Leucyl-tRNA synthetase, METHIONINE | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
2AJI
 
 | | Crystal structure of the editing domain of E. coli leucyl-tRNA synthetase complexes with isoleucine | | Descriptor: | ISOLEUCINE, Leucyl-tRNA synthetase | | Authors: | Liu, Y, Liao, J, Zhu, B, Wang, E.D, Ding, J. | | Deposit date: | 2005-08-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination
Biochem.J., 394, 2006
|
|
5J5C
 
 | | Crystal structure of ARL1-GTP and DCB domain of BIG1 complex | | Descriptor: | ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, R, Wang, Z, Zhang, T, Ding, J. | | Deposit date: | 2016-04-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for targeting BIG1 to Golgi apparatus through interaction of its DCB domain with Arl1
J Mol Cell Biol, 2016
|
|
1NLN
 
 | | CRYSTAL STRUCTURE OF HUMAN ADENOVIRUS 2 PROTEINASE WITH ITS 11 AMINO ACID COFACTOR AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, Adenain, PVIC | | Authors: | McGrath, W.J, Ding, J, Sweet, R.M, Mangel, W.F. | | Deposit date: | 2003-01-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold
Biochim.Biophys.Acta, 1648, 2003
|
|
1HQU
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, POL POLYPROTEIN | | Authors: | Hsiou, Y, Ding, J, Arnold, E. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
1L3K
 
 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
5H4B
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
1HYS
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH A POLYPURINE TRACT RNA:DNA | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*AP*AP*AP*AP*AP*GP*TP*GP*GP*CP*TP*G)-3', 5'-R(*UP*CP*AP*GP*CP*CP*AP*CP*UP*UP*UP*UP*UP*AP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', FAB-28 MONOCLONAL ANTIBODY FRAGMENT HEAVY CHAIN, ... | | Authors: | Sarafianos, S.G, Das, K, Tantillo, C, Clark Jr, A.D, Ding, J, Whitcomb, J, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2001-01-22 | | Release date: | 2001-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA.
EMBO J., 20, 2001
|
|
1QE1
 
 | | CRYSTAL STRUCTURE OF 3TC-RESISTANT M184I MUTANT OF HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | REVERSE TRANSCRIPTASE, SUBUNIT P51, SUBUNIT P66 | | Authors: | Sarafianos, S.G, Das, K, Ding, J, Hughes, S.H, Arnold, E. | | Deposit date: | 1999-07-12 | | Release date: | 1999-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3IU3
 
 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
4FGB
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I apo form | | Descriptor: | Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
4FG9
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-320 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
4FG8
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-315 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
4FG7
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-293 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
2AKE
 
 | | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Shen, N, Guo, L, Yang, B, Jin, Y, Ding, J. | | Deposit date: | 2005-08-03 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) reveals the molecular basis of tRNA recognition and specificity
Nucleic Acids Res., 34, 2006
|
|
2DSB
 
 | |
2DSC
 
 | |
2DSD
 
 | |
2DR2
 
 | | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Shen, N, Guo, L, Yang, B, Jin, Y, Ding, J. | | Deposit date: | 2006-06-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) reveals the molecular basis of tRNA recognition and specificity
Nucleic Acids Res., 34, 2006
|
|
1YZX
 
 | | Crystal structure of human kappa class glutathione transferase | | Descriptor: | Glutathione S-transferase kappa 1, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE | | Authors: | Li, J, Xia, Z, Ding, J. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Thioredoxin-like domain of human kappa class glutathione transferase reveals sequence homology and structure similarity to the theta class enzyme
PROTEIN SCI., 14, 2005
|
|
8JDB
 
 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyoctanoic acid | | Descriptor: | (2R)-2-oxidanyloctanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDD
 
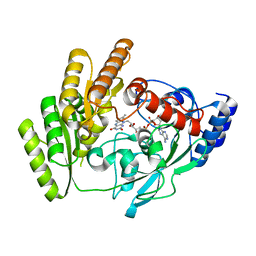 | | Crystal structure of H405A mLDHD in apo form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, mitochondrial | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDT
 
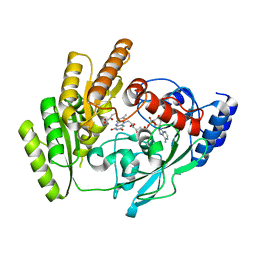 | | Crystal structure of mLDHD in complex with 2-ketobutanoic acid | | Descriptor: | 2-KETOBUTYRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDG
 
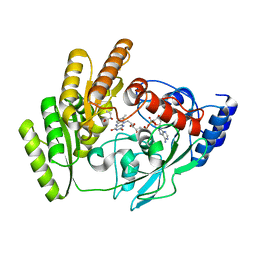 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxybutanoic acid | | Descriptor: | (2R)-2-oxidanylbutanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDQ
 
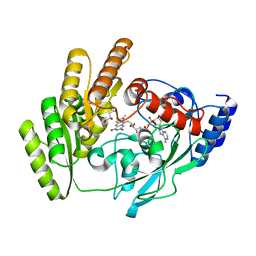 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyisocaproic acid | | Descriptor: | (2R)-2-hydroxy-4-methylpentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
