4FTH
 
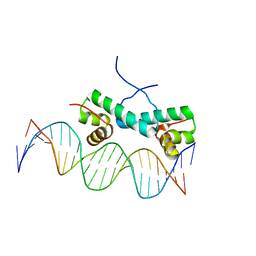 | | Crystal Structure of NtrC4 DNA-binding domain bound to double-stranded DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*GP*CP*AP*T)-3', 5'-D(P*GP*AP*TP*GP*CP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*A)-3', Transcriptional regulator (NtrC family) | | Authors: | Vidangos, N.K, Heideker, J, Lyubimov, A.Y, Lamers, M, Huo, Y, Pelton, J.G, Ton, J, Gralla, J.D, Kuriyan, J, Berger, J.M, Wemmer, D.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | DNA Recognition by a sigma (54) Transcriptional Activator from Aquifex aeolicus.
J.Mol.Biol., 426, 2014
|
|
9EO6
 
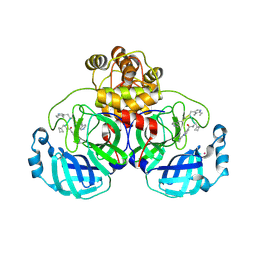 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL11, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9EOX
 
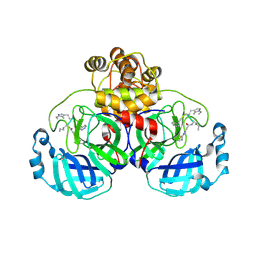 | | SARS-CoV2 major protease in covalent complex with a soluble inhibitor. | | Descriptor: | 3C-like proteinase nsp5, POTASSIUM ION, Soluble inhibitor | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9EOR
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL12. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL12, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
2VHR
 
 | |
2WBZ
 
 | |
2VI1
 
 | |
2VI4
 
 | |
2VHK
 
 | |
2VI3
 
 | |
2VI2
 
 | |
2VU6
 
 | |
2VU7
 
 | | Atomic resolution (1.08 A) structure of purified thaumatin I grown in sodium meso-tartrate at 4 C | | Descriptor: | 1,2-ETHANEDIOL, S,R MESO-TARTARIC ACID, Thaumatin-1 | | Authors: | Jakoncic, J, Asherie, N, Ginsberg, C. | | Deposit date: | 2008-05-21 | | Release date: | 2009-07-14 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Tartrate Chirality Determines Thaumatin Crystal Habit
Cryst.Growth Des., 9, 2009
|
|
2LAI
 
 | |
4WL0
 
 | |
5ONI
 
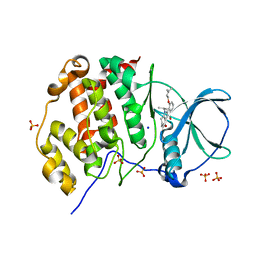 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
6IC4
 
 | |
4U1R
 
 | |
6NUJ
 
 | |
4XZ2
 
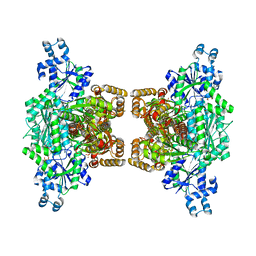 | | Human platelet phosphofructokinase in an R-state in complex with ADP and F6P, crystal form I | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kloos, M, Strater, N. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of human platelet phosphofructokinase-1 locked in an activated conformation.
Biochem.J., 469, 2015
|
|
6U5R
 
 | | Calcium-bound MthK closed state | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel MthK | | Authors: | Fan, C, Nimigean, C.M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
6U6H
 
 | | Calcium-bound MthK open-inactivated state 3 | | Descriptor: | Calcium-gated potassium channel MthK | | Authors: | Chen, F, Crina, N. | | Deposit date: | 2019-08-29 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
6U5P
 
 | | Calcium-bound MthK gating ring state 1 | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel MthK | | Authors: | Fan, C, Nimigean, C.M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
6U68
 
 | |
8G8C
 
 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
