2J5P
 
 | | E. coli FtsK gamma domain | | Descriptor: | DNA TRANSLOCASE FTSK | | Authors: | Sivanathan, V, Allen, M.D, de Bekker, C, Baker, R, Arciszewska, L, Freund, S.M, Bycroft, M, Lowe, J, Sherratt, D.J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ftsk Gamma Domain Directs Oriented DNA Translocation by Interacting with Kops.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2J5O
 
 | | Pseudomonas aeruginosa FtsK gamma domain | | Descriptor: | DNA TRANSLOCASE FTSK | | Authors: | Sivanathan, V, Allen, M.D, deBekker, C, Baker, R, Arciszewska, L, Freund, S.M, Bycroft, M, Lowe, J, Sherratt, D.J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ftsk Gamma Domain Directs Oriented DNA Translocation by Interacting with Kops.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1UOL
 
 | |
1EBD
 
 | | DIHYDROLIPOAMIDE DEHYDROGENASE COMPLEXED WITH THE BINDING DOMAIN OF THE DIHYDROLIPOAMIDE ACETYLASE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE, DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mande, S.S, Sarfaty, S, Allen, M.D, Perham, R.N, Hol, W.G.J. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase.
Structure, 4, 1996
|
|
1H3Z
 
 | |
2VYR
 
 | | Structure of human MDM4 N-terminal domain bound to a single domain antibody | | Descriptor: | HUMAN SINGLE DOMAIN ANTIBODY, MDM4 PROTEIN, SULFATE ION | | Authors: | Yu, G.W, Vaysburd, M, Allen, M.D, Settanni, G, Fersht, A.R. | | Deposit date: | 2008-07-28 | | Release date: | 2008-11-25 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human Mdm4 N-Terminal Domain Bound to a Single-Domain Antibody.
J.Mol.Biol., 385, 2009
|
|
2VE9
 
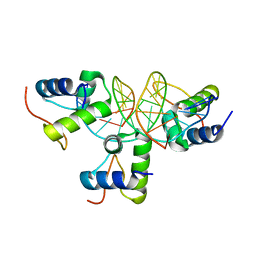 | | Xray structure of KOPS bound gamma domain of FtsK (P. aeruginosa) | | Descriptor: | 5'-D(*AP*CP*CP*AP*GP*GP*GP*CP*AP*GP *GP*GP*CP*GP*AP*C)-3', 5'-D(*GP*TP*CP*GP*CP*CP*CP*TP*GP*CP *CP*CP*TP*GP*GP*T)-3', DNA TRANSLOCASE FTSK, ... | | Authors: | Lowe, J, Allen, M.D, Sherratt, D.J. | | Deposit date: | 2007-10-17 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of Sequence-Directed DNA Loading and Translocation by Ftsk.
Mol.Cell, 31, 2008
|
|
2W0T
 
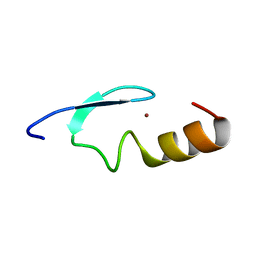 | |
2WZO
 
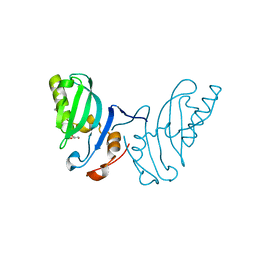 | | The structure of the FYR domain | | Descriptor: | GLYCEROL, TRANSFORMING GROWTH FACTOR BETA REGULATOR 1 | | Authors: | Garcia-Alai, M.M, Allen, M.D, Joerger, A.C, Bycroft, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Fyr Domain of Transforming Growth Factor Beta Regulator 1 (Tbrg1)(Pna)
Protein Sci., 19, 2010
|
|
2X35
 
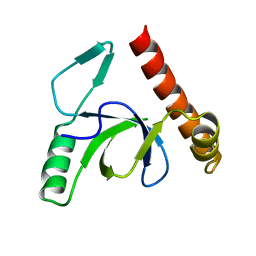 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | PEREGRIN | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2X4W
 
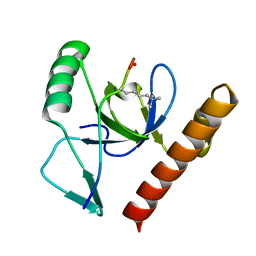 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | FORMIC ACID, HISTONE H3.2, PEREGRIN | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2X4Y
 
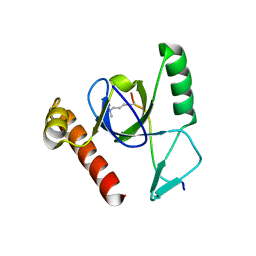 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | HISTONE H3.2, PEREGRIN, SULFATE ION | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2VYT
 
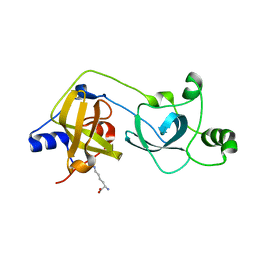 | | The MBT repeats of human SCML2 bind to peptides containing mono methylated lysine. | | Descriptor: | N-METHYL-LYSINE, SEX COMB ON MIDLEG-LIKE PROTEIN 2, TRIETHYLENE GLYCOL | | Authors: | Santiveri, C.M, Lechtenberg, B.C, Allen, M.D, Sathyamurthy, A, Jaulent, A.M, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2008-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Malignant Brain Tumor Repeats of Human Scml2 Bind to Peptides Containing Monomethylated Lysine.
J.Mol.Biol., 382, 2008
|
|
2X4X
 
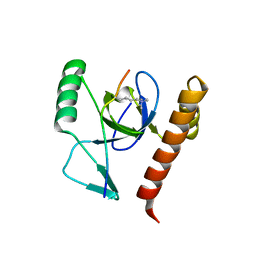 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | HISTONE H3.2, PEREGRIN, SULFATE ION | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WXC
 
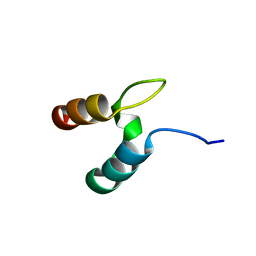 | | The folding mechanism of BBL: Plasticity of transition-state structure observed within an ultrafast folding protein family. | | Descriptor: | DIHYDROLIPOYLTRANSSUCCINASE | | Authors: | Neuweiler, H, Sharpe, T.D, Rutherford, T.J, Johnson, C.M, Allen, M.D, Ferguson, N, Fersht, A.R. | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Folding Mechanism of Bbl: Plasticity of Transition-State Structure Observed within an Ultrafast Folding Protein Family.
J.Mol.Biol., 390, 2009
|
|
4CYI
 
 | | Chaetomium thermophilum Pan3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
4CYK
 
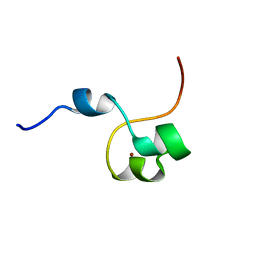 | | Structural basis for binding of Pan3 to Pan2 and its function in mRNA recruitment and deadenylation | | Descriptor: | PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3, ZINC ION | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation.
Embo J., 33, 2014
|
|
4CYJ
 
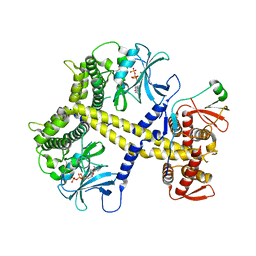 | | Chaetomium thermophilum Pan2:Pan3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
1OI1
 
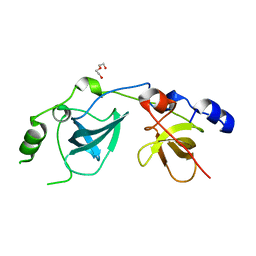 | | Crystal Structure of the MBT domains of Human SCML2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, SCML2 PROTEIN | | Authors: | Sathyamurthy, A, Allen, M.D, Murzin, A.G, Bycroft, M. | | Deposit date: | 2003-06-04 | | Release date: | 2003-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Malignant Brain Tumor (Mbt) Repeats in Sex Comb on Midleg-Like 2 (Scml2).
J.Biol.Chem., 278, 2003
|
|
1OI0
 
 | | CRYSTAL STRUCTURE OF AF2198, A JAB1/MPN DOMAIN PROTEIN FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | HYPOTHETICAL PROTEIN AF2198, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ZINC ION | | Authors: | Tran, H.J.T.T, Allen, M.D, Lowe, J, Bycroft, M. | | Deposit date: | 2003-06-04 | | Release date: | 2003-08-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Jab1/Mpn Domain and its Implications for Proteasome Function
Biochemistry, 42, 2003
|
|
8P8Z
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 1963 | | Descriptor: | 3-[3-cyano-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 1963
To Be Published
|
|
8PA3
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2500 | | Descriptor: | 7-[(1~{S})-1-[4-(2-azanylethyl)phenoxy]ethyl]-3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2500
To Be Published
|
|
8P93
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2163 | | Descriptor: | 3-[3-fluoranyl-4-[(piperidin-4-ylsulfonylamino)methyl]phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2163
To Be Published
|
|
8P9Q
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2455 | | Descriptor: | 7-[(1~{S})-1-acetyloxyethyl]-3-[3-fluoranyl-4-(sulfamoylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2455
To Be Published
|
|
8PAA
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2552 | | Descriptor: | 7-[(1~{S})-1-[4-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]phenyl]carbonyloxyethyl]-3-[6-(morpholin-4-ylmethyl)pyridin-3-yl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2552
To Be Published
|
|
