4O66
 
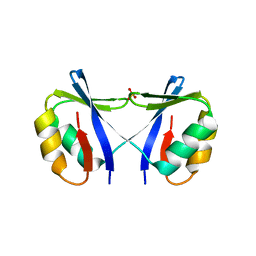 | | Crystal Structure of SMARCAL1 HARP substrate recognition domain | | Descriptor: | SODIUM ION, SULFATE ION, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 | | Authors: | Mason, A.C, Eichman, B.F. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structure-specific nucleic acid-binding domain conserved among DNA repair proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7MBH
 
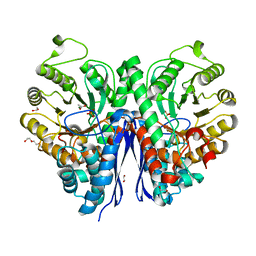 | | Structure of Human Enolase 2 in complex with phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Gamma-enolase, ... | | Authors: | Leonard, P.G, Hicks, K.G, Rutter, J. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase.
Science, 379, 2023
|
|
7NFW
 
 | |
3RKY
 
 | |
3RN5
 
 | | Structural basis of cytosolic DNA recognition by innate immune receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*AP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*TP*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*G)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RNU
 
 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
7NDY
 
 | | Di-phosphorylated Barrier-to-Autointegration Factor (BAF) in complex with LEM domain of Emerin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Barrier-to-autointegration factor, N-terminally processed, ... | | Authors: | Marcelot, A, Le Du, M.H, Hoffmann, G, Zinn-Justin, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
3R5J
 
 | |
7LHZ
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | | Descriptor: | (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*AP*TP*CP*AP*TP*AP*CP*AP*AP*CP*GP*TP*AP*A)-3'), ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2021-01-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
3RBM
 
 | | Crystal structure of Plasmodium vivax geranylgeranylpyrophosphate synthase complexed with BPH -703 | | Descriptor: | 3-(2,2-diphosphonoethyl)-1-dodecyl-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Liu, Y.L, No, J.H, Oldfield, E. | | Deposit date: | 2011-03-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Lipophilic analogs of zoledronate and risedronate inhibit Plasmodium geranylgeranyl diphosphate synthase (GGPPS) and exhibit potent antimalarial activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7LZ4
 
 | | Crystal structure of A211D mutant of Protein Kinase A RIa subunit, a Carney Complex mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed | | Authors: | Del Rio, J, Wu, J, Taylor, S.S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.155 Å) | | Cite: | Noncanonical protein kinase A activation by oligomerization of regulatory subunits as revealed by inherited Carney complex mutations.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LW6
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with Raltegravir | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, White, S.W, Rankovik, Z. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
3SGV
 
 | |
3S21
 
 | | Crystal structure of cerulenin bound Xanthomonas campestri OleA (co-crystal) | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-05-16 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7001 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
7M6B
 
 | | The Crystal Structure of Mcbe1 | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2021-03-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
3SE8
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC03 in complex with HIV-1 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
7M3I
 
 | |
3SFY
 
 | |
4NMM
 
 | | Crystal Structure of a G12C Oncogenic Variant of Human KRas Bound to a Novel GDP Competitive Covalent Inhibitor | | Descriptor: | 5'-O-[(S)-{[(S)-[2-(acetylamino)ethoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]guanosine, GTPase KRas, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Lim, S.M, Westover, K.D. | | Deposit date: | 2013-11-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3S41
 
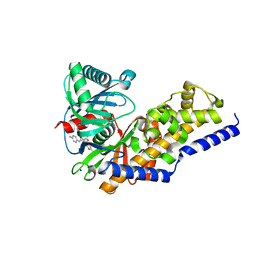 | | Glucokinase in complex with activator and glucose | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2011-05-18 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Designing glucokinase activators with reduced hypoglycemia risk: discovery of N,N-dimethyl-5-(2-methyl-6-((5-methylpyrazin-2-yl)-carbamoyl)benzofuran-4-yloxy)pyrimidine-2-carboxamide as a clinical candidate for the treatment of type 2 diabetes mellitus
MEDCHEMCOMM, 2, 2011
|
|
4NSE
 
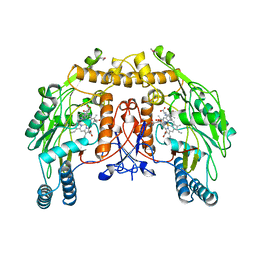 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, L-ARG COMPLEX | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1998-10-07 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
4NWK
 
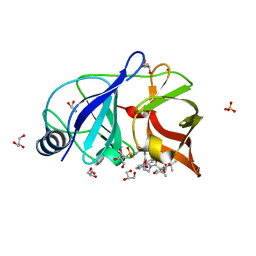 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | Descriptor: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4NK2
 
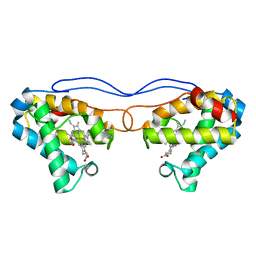 | | Crystal structure of Hell's gate globin IV | | Descriptor: | Hemoglobin-like protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jamil, F. | | Deposit date: | 2013-11-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of truncated haemoglobin from an extremely thermophilic and acidophilic bacterium.
J.Biochem., 156, 2014
|
|
7UN7
 
 | |
4NWL
 
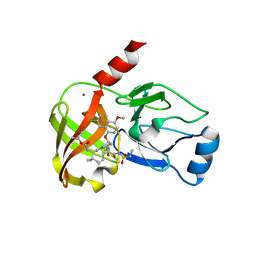 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-650032 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-4-((7-chloro-4-methoxy-1-isoquinolinyl)o xy)-n-((1r,2s)-1-((cyclopropylsulfonyl)carbamoyl)-2-vinylc yclopropyl)-l-prolinamide | | Descriptor: | HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, ZINC ION | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
