6KKT
 
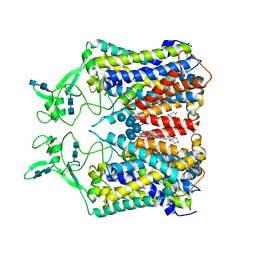 | | human KCC1 structure determined in KCl and lipid nanodisc | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6KKR
 
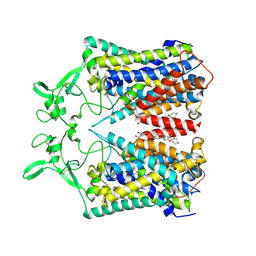 | | human KCC1 structure determined in KCl and detergent GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6MO5
 
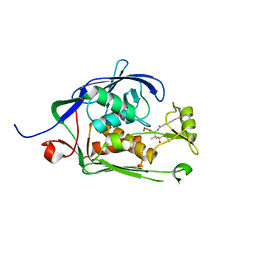 | | Co-Crystal structure of P. aeruginosa LpxC-50228 complex | | Descriptor: | MAGNESIUM ION, N-[(2S)-1-(hydroxyamino)-3-methyl-3-{[(oxetan-3-yl)methyl]sulfonyl}-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
7WRA
 
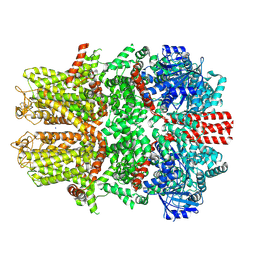 | | Mouse TRPM8 in LMNG in ligand-free state | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
7WRD
 
 | |
7WRF
 
 | | Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Icilin, SODIUM ION, ... | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
7WRB
 
 | |
7WRC
 
 | | Mouse TRPM8 in LMNG in the presence of calcium, icilin and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Icilin, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
7WRE
 
 | |
6KKU
 
 | | human KCC1 structure determined in NaCl and GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6MOO
 
 | | Co-Crystal structure of P. aeruginosa LpxC-achn975 complex | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MO4
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOD
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50432 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-[(1S)-2-(hydroxyamino)-1-(3-methoxy-1,1-dioxo-1lambda~6~-thietan-3-yl)-2-oxoethyl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, ... | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
1ASW
 
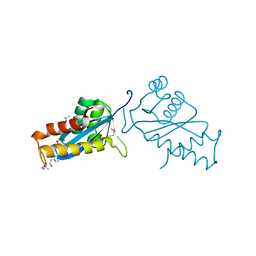 | | AVIAN SARCOMA VIRUS INTEGRASE CATALYTIC CORE DOMAIN CRYSTALLIZED FROM 20% PEG 4000, 10% ISOPROPANOL, HEPES PH 7.5 USING SELENOMETHIONINE SUBSTITUTED PROTEIN; DATA COLLECTED AT-165 DEGREES C | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE, ISOPROPYL ALCOHOL | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-08-25 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure of the catalytic domain of avian sarcoma virus integrase.
J.Mol.Biol., 253, 1995
|
|
5AK7
 
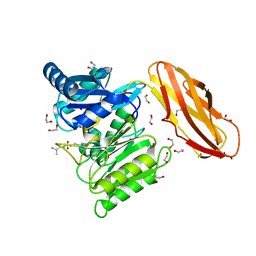 | | Structure of wt Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
7FAY
 
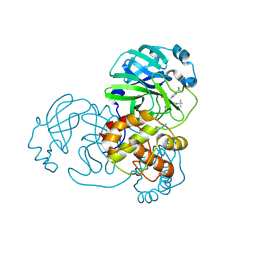 | | Crystal structure of SARS-CoV-2 main protease in complex with (R)-1a | | Descriptor: | (2~{R})-~{N}-[(1~{R})-2-(~{tert}-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-~{N}-(4-~{tert}-butylphenyl)-2-oxidanyl-propanamide, 3C-like proteinase | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
7FAZ
 
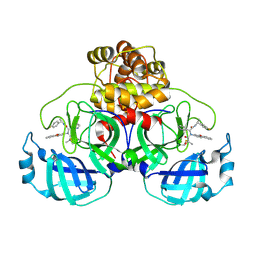 | | Crystal structure of the SARS-CoV-2 main protease in complex with Y180 | | Descriptor: | (2~{R})-~{N}-dibenzofuran-3-yl-~{N}-[(1~{R})-2-[[(1~{S})-1-(4-fluorophenyl)ethyl]amino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]-2-oxidanyl-propanamide, 3C-like proteinase, SODIUM ION | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
5AK8
 
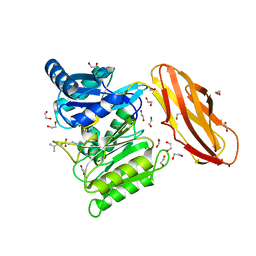 | | Structure of C351A mutant of Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
2HX4
 
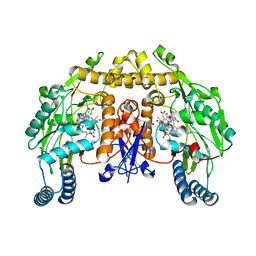 | | Rat nNOS heme domain complexed with 4-N-(Nw-nitro-L-argininyl)-trans-4-hydroxyamino-L-proline amide | | Descriptor: | (4R)-4-(HYDROXY{N~5~-[IMINO(NITROAMINO)METHYL]-L-ORNITHYL}AMINO)-L-PROLINAMIDE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX2
 
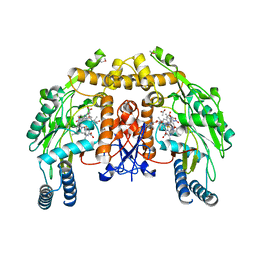 | | Bovine eNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | Descriptor: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX3
 
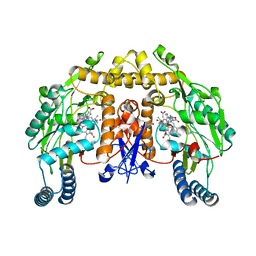 | | Rat nNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | Descriptor: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
1ITX
 
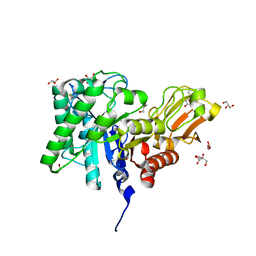 | | Catalytic Domain of Chitinase A1 from Bacillus circulans WL-12 | | Descriptor: | GLYCEROL, Glycosyl Hydrolase | | Authors: | Iwahori, F, Matsumoto, T, Watanabe, T, Nonaka, T. | | Deposit date: | 2002-02-13 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Three-dimensional structure of the catalytic domain of chitinase A1 from Bacillus circulans WL-12 at a very high resolution
PROC.JPN.ACAD.,SER.B, 75, 1999
|
|
4NB6
 
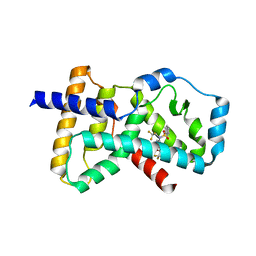 | | Crystal structure of the ligand binding domain of RORC with T0901317 | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear receptor ROR-gamma | | Authors: | Hymowitz, S.G, Boenig-de Leon, G. | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of substituted hexafluoroisopropanol-arylsulfonamides as modulators of RORc.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6L3R
 
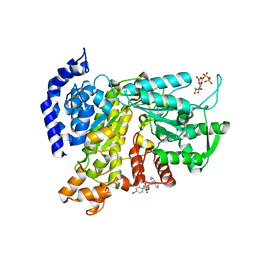 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide | | Descriptor: | 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
6L7L
 
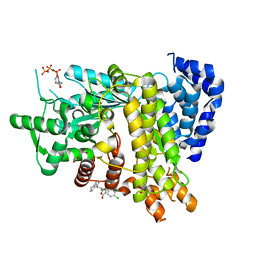 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide | | Descriptor: | 5-chloro-2-(N-((1S,2R)-2-(2,3-dihydro-1H-inden-4-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)sulfamoyl)benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
