6XGX
 
 | | ISCth4 transposase, strand transfer complex 1, STC1 | | Descriptor: | DNA (21-MER), DNA (25-MER), DNA (47-MER), ... | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
6HAB
 
 | | Crystal structure of BiP V461F (apo) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
1HTO
 
 | |
6HB1
 
 | | Structure of Hgh1, crystal form I | | Descriptor: | CHLORIDE ION, Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
1HYV
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH TETRAPHENYL ARSONIUM | | Descriptor: | CHLORIDE ION, INTEGRASE, SULFATE ION, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HIB
 
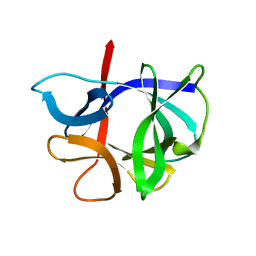 | | THE STRUCTURE OF AN INTERLEUKIN-1 BETA MUTANT WITH REDUCED BIOACTIVITY SHOWS MULTIPLE SUBTLE CHANGES IN CONFORMATION THAT AFFECT PROTEIN-PROTEIN RECOGNITION | | Descriptor: | INTERLEUKIN-1 BETA | | Authors: | Camacho, N.P, Smith, D.R, Goldman, A, Schneider, B, Green, D, Young, P.R, Berman, H.M. | | Deposit date: | 1993-03-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an interleukin-1 beta mutant with reduced bioactivity shows multiple subtle changes in conformation that affect protein-protein recognition.
Biochemistry, 32, 1993
|
|
6HCI
 
 | |
1HYZ
 
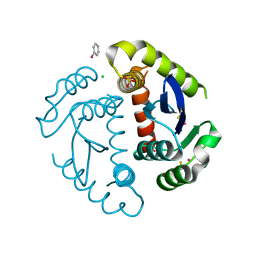 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH A DERIVATIVE OF TETRAPHENYL ARSONIUM. | | Descriptor: | (3,4-DIHYDROXY-PHENYL)-TRIPHENYL-ARSONIUM, CHLORIDE ION, INTEGRASE, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HYN
 
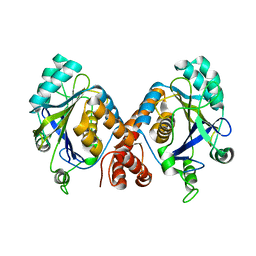 | |
6HN3
 
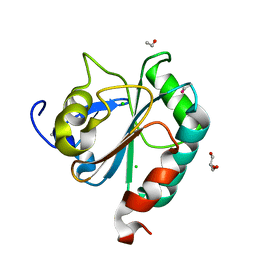 | | wildtype form (apo) of human GPX4 with Se-Cys46 | | Descriptor: | CHLORIDE ION, ETHANOL, GLYCEROL, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Hoffmann, J, Schnirch, L, Eaton, J.K, Badock, V, Gradl, S. | | Deposit date: | 2018-09-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Crystal structures of the selenoprotein glutathione peroxidase 4 in its apo form and in complex with the covalently bound inhibitor ML162.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6HZZ
 
 | | Structure of human D-glucuronyl C5 epimerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HK4
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C22 | | Descriptor: | 3-azanyl-6-(4-morpholin-4-ylsulfonylphenyl)-~{N}-pyridin-3-yl-pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLY-SER-HIS-GLY-HIS-HIS-HIS-HIS-HIS, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
1HZ5
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS, WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6XTA
 
 | | Recombinant human butyrylcholinesterase in complex with (2S)-N-[2-(1-benzylazepan-4-yl)ethyl]-2-(butylamino)-3-(1H-indol-3-yl)propanamide | | Descriptor: | (2~{S})-2-(butylamino)-3-(1~{H}-indol-3-yl)-~{N}-[2-[(4~{R})-1-(phenylmethyl)azepan-4-yl]ethyl]propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Nachon, F, Meden, A, Knez, D, Gobec, S. | | Deposit date: | 2020-01-15 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-activity relationship study of tryptophan-based butyrylcholinesterase inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
1HZ6
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HZ9
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
6HB2
 
 | | Structure of Hgh1, crystal form I, Selenomethionine derivative | | Descriptor: | CHLORIDE ION, Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
1HZA
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HQD
 
 | | PSEUDOMONAS CEPACIA LIPASE COMPLEXED WITH TRANSITION STATE ANALOGUE OF 1-PHENOXY-2-ACETOXY BUTANE | | Descriptor: | (RP,SP)-O-(2R)-(1-PHENOXYBUT-2-YL)-METHYLPHOSPHONIC ACID CHLORIDE, CALCIUM ION, LIPASE | | Authors: | Luic, M, Tomic, S, Lescic, I, Ljubovic, E, Sepac, D, Sunjic, V, Vitale, L, Saenger, W, Kojic-Prodic, B. | | Deposit date: | 2000-12-15 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex of Burkholderia cepacia lipase with transition state analogue of 1-phenoxy-2-acetoxybutane: biocatalytic, structural and modelling study.
Eur.J.Biochem., 268, 2001
|
|
1HZB
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZC
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
6I7F
 
 | | Dehaloperoxidase B from Amphitrite ornata - complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moreno-Chicano, T, Ebrahim, A.E, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6I7C
 
 | | Dye type peroxidase Aa from Streptomyces lividans: imidazole complex | | Descriptor: | Deferrochelatase/peroxidase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno-Chicano, T, Ebrahim, A.E, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6DNK
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-06-06 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
4ZL1
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18X at 1.86 A resolution | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Huang, J, Wu, D, Ouyang, P, Lu, W, Pu, J. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18XYW at 1.86 A resolution
To Be Published
|
|
