3KKW
 
 | | Crystal structure of His-tagged form of PA4794 protein | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, SULFATE ION | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Double trouble-Buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments.
Protein Sci., 23, 2014
|
|
3KRV
 
 | | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rakonjac, N, Rezacova, P, Borek, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity
To be Published
|
|
3I4Q
 
 | | Structure of a putative inorganic pyrophosphatase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | APC40078, SODIUM ION | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3OWC
 
 | | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, Probable acetyltransferase | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa
To be Published
|
|
3LL7
 
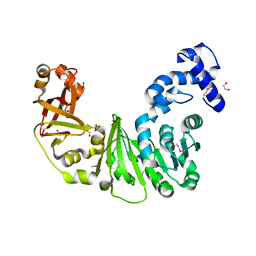 | | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase | | Authors: | Chang, C, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83
To be Published
|
|
3L9A
 
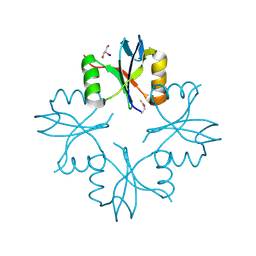 | | Structure of the C-terminal domain from a Streptococcus mutans hypothetical | | Descriptor: | GLYCEROL, SODIUM ION, uncharacterized protein | | Authors: | Singer, A.U, Cuff, M.E, Xu, X, Cui, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-04 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the C-terminal domain from a Streptococcus mutans hypothetical
To be Published
|
|
3LDU
 
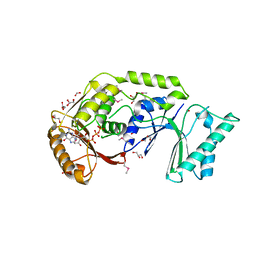 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
3LFT
 
 | |
3IR9
 
 | |
4R5Q
 
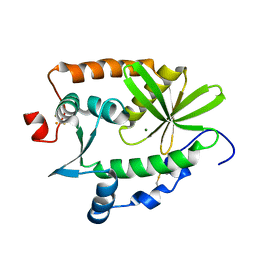 | | Crystal structure and nuclease activity of the CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis containing a [2Fe-2S] cluster | | Descriptor: | CRISPR-associated exonuclease, Cas4 family, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nocek, B, Skarina, T, Lemak, S, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure and nuclease activity of the CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis containing a [2Fe-2S] cluster
To be Published
|
|
3LM9
 
 | | Crystal structure of fructokinase with ADP and Fructose bound in the active site | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ZINC ION, ... | | Authors: | Nocek, B, Stein, A, Cuff, M, Volkart, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
3ICY
 
 | |
3LEQ
 
 | | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A | | Descriptor: | uncharacterized protein cvnB5 | | Authors: | Stein, A.J, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A
To be Published
|
|
1M6Y
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | Authors: | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-07-17 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
3IBZ
 
 | | Crystal structure of putative tellurium resistant like protein (TerD) from Streptomyces coelicolor A3(2) | | Descriptor: | CALCIUM ION, Putative tellurium resistant like protein TerD, SULFATE ION | | Authors: | Klimecka, M, Chruszcz, M, Cymborowski, M, Xu, X, Cui, H, Joachimiak, A, Edwards, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of putative tellurium resistant like protein (TerD) from Streptomyces coelicolor A3(2)
To be Published
|
|
4Z7X
 
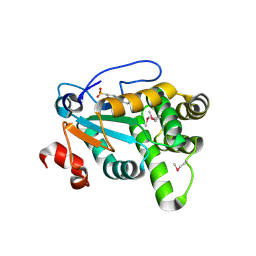 | | MdbA protein, a thiol-disulfide oxidoreductase from Actinomyces oris. | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, MdbA | | Authors: | OSIPIUK, J, Reardon-Robinson, M.E, Ton-That, H, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Disulfide Bond-forming Machine Is Linked to the Sortase-mediated Pilus Assembly Pathway in the Gram-positive Bacterium Actinomyces oris.
J.Biol.Chem., 290, 2015
|
|
3LUP
 
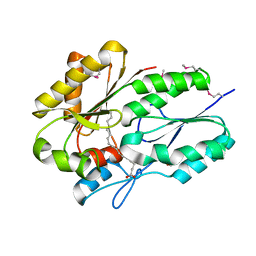 | | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae | | Descriptor: | 9-OCTADECENOIC ACID, DegV family protein, GLYCEROL | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae
To be Published
|
|
3B49
 
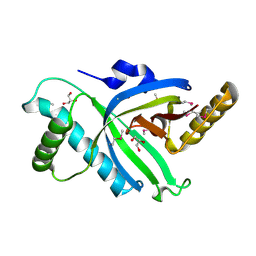 | |
3FYN
 
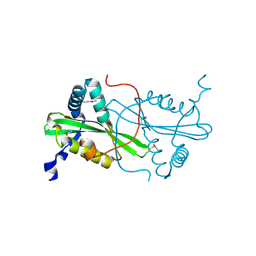 | | Crystal structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3 | | Descriptor: | ACETATE ION, Integron gene cassette protein HFX_CASS3, MAGNESIUM ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3
To be Published
|
|
3LG2
 
 | | A Ykr043C/ fructose-1,6-bisphosphate product complex following ligand soaking | | Descriptor: | PHOSPHATE ION, Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3LVU
 
 | | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ABC transporter, ... | | Authors: | Chang, C, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi
To be Published
|
|
3LXQ
 
 | | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A | | Descriptor: | CHLORIDE ION, Uncharacterized protein VP1736 | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A
To be Published
|
|
4R1G
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cloxacillin | | Descriptor: | CLOXACILLIN (OPEN FORM), Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cloxacillin
To be Published
|
|
4GYM
 
 | | Crystal structure of Glyoxalase/bleomycin resistance protein/dioxygenase from Conexibacter woesei DSM 14684 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, POTASSIUM ION, ... | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of Glyoxalase/bleomycin resistance protein/dioxygenase from Conexibacter woesei DSM 14684
To be Published
|
|
1XAF
 
 | | Crystal Structure of Protein of Unknown Function YfiH from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, GLYCEROL, ZINC ION, ... | | Authors: | Kim, Y, Maltseva, N, Dementieva, I, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-25 | | Release date: | 2004-08-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution.
Proteins, 63, 2006
|
|
