8JP2
 
 | | Crystal structure of AKR1C1 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8JP1
 
 | | Crystal structure of AKR1C3 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
7WVP
 
 | |
7WVQ
 
 | |
2L65
 
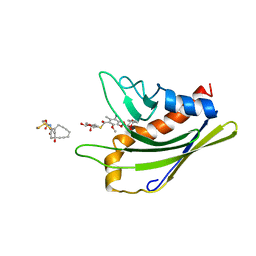 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
3VH7
 
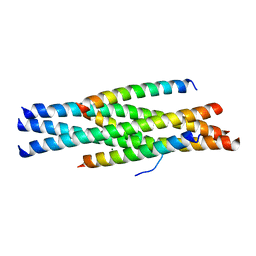 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P21 | | Descriptor: | CP32M, Envelope glycoprotein gp160, MAGNESIUM ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
9IMO
 
 | | Crystal structure of Tubulin-RB3-TTL-Y12 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-04 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IM5
 
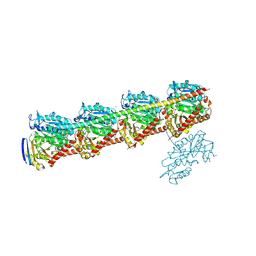 | | Tubulin-RB3(MUT)-TTL-Y12 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-02 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7MEJ
 
 | |
7MDW
 
 | |
7ME7
 
 | |
7MTR
 
 | | CryoEM Structure of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethyl)thieno[2,3-d]pyrimidin-4-amine, GLUTAMIC ACID, ... | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
7MTQ
 
 | | CryoEM Structure of Full-Length mGlu2 in Inactive-State Bound to Antagonist LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
7MTS
 
 | | CryoEM Structure of mGlu2 - Gi Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethyl)thieno[2,3-d]pyrimidin-4-amine, GLUTAMIC ACID, ... | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
7N9T
 
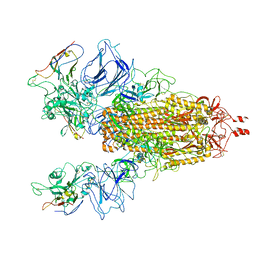 | | CryoEM structure of SARS-CoV-2 Spike in complex with Nb17 | | Descriptor: | Nanobody Nb17, Spike glycoprotein | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2021-06-18 | | Release date: | 2021-08-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes
Nat Commun, 12, 2021
|
|
3VIE
 
 | | HIV-gp41 fusion inhibitor Sifuvirtide | | Descriptor: | Envelope glycoprotein gp160, Sifuvirtide | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-09-29 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide
J.Biol.Chem., 287, 2012
|
|
9JSB
 
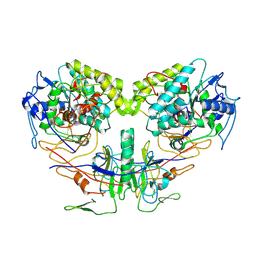 | | guide-bound NbaSPARDA complexes | | Descriptor: | Ago, DREN-APAZ, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-09-30 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
9JSP
 
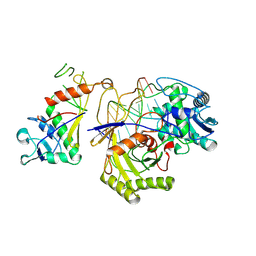 | | inactive NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(P*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DREN-APAZ, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-09-30 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
9JT2
 
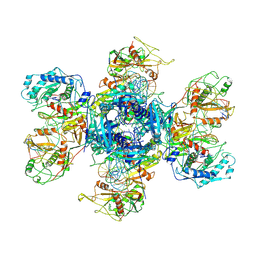 | | substrate-bound NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(*TP*AP*TP*CP*GP*TP*CP*AP*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DNA (5'-D(P*GP*AP*TP*AP*CP*TP*AP*C)-3'), ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-10-01 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
5XP5
 
 | | C-Src in complex with ATP-Chf | | Descriptor: | MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[(S)-fluoranyl-[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Duan, Y, Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
9JSZ
 
 | | active NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(*TP*AP*TP*CP*GP*TP*CP*AP*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DREN-APAZ, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-10-01 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
3VGY
 
 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P321 | | Descriptor: | CP32M, Envelope glycoprotein gp160, SULFATE ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
8WCE
 
 | | Cryo-EM structure of a protein-RNA complex | | Descriptor: | MAGNESIUM ION, RNA (31-MER), RNA (66-MER), ... | | Authors: | Li, Z. | | Deposit date: | 2023-09-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Molecular mechanism for target RNA recognition and cleavage of Cas13h.
Nucleic Acids Res., 52, 2024
|
|
8WD8
 
 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
7BPI
 
 | | The crystal structue of PDE10A complexed with 14 | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2020-03-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4000864 Å) | | Cite: | Discovery of highly selective and orally available benzimidazole-based phosphodiesterase 10 inhibitors with improved solubility and pharmacokinetic properties for treatment of pulmonary arterial hypertension.
Acta Pharm Sin B, 10, 2020
|
|
