6MHC
 
 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 37 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MN7
 
 | | Cryo-EM structure of BG505.SOSIP.664 in complex with BF520.1 antigen binding fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BF520.1 Fab variable region, ... | | Authors: | Williams, J.A, Lee, K.K, Overbaugh, J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-03-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kappa chain maturation helps drive rapid development of an infant HIV-1 broadly neutralizing antibody lineage.
Nat Commun, 10, 2019
|
|
6MS3
 
 | | Crystal structure of the GH43 protein BlXynB mutant (K247S) from Bacillus licheniformis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside Hydrolase Family 43, ... | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
6MXF
 
 | | MicroED structure of thiostrepton at 1.9 A resolution | | Descriptor: | Thiostrepton | | Authors: | Jones, C.G, Martynowycz, M.W, Hattne, J, Fulton, T, Stoltz, B.M, Rodriguez, J.A, Nelson, H.M, Gonen, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination.
ACS Cent Sci, 4, 2018
|
|
6N3Z
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 4 | | Descriptor: | Epoxide hydrolase TrEH, N-methyl-4-{[trans-4-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)cyclohexyl]oxy}benzamide | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6MR6
 
 | |
6N5H
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 5 | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriano, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors
Int. J. Biol. Macromol., 129, 2019
|
|
6MWN
 
 | |
6N04
 
 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
6N5F
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 3 | | Descriptor: | Epoxide hydrolase TrEH, N-(8-amino-8-oxooctyl)nonanamide | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5G
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
7JJP
 
 | | Sheep Connexin-50 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
2WSA
 
 | | Crystal Structure of Leishmania major N-myristoyltransferase (NMT) with bound myristoyl-CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | N-Myristoyltransferase Inhibitors as New Leads to Treat Sleeping Sickness.
Nature, 464, 2010
|
|
7JMD
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JLW
 
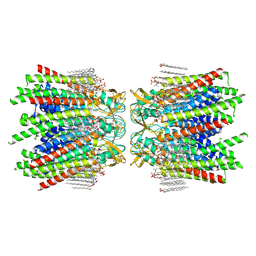 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
5IJT
 
 | |
7JMC
 
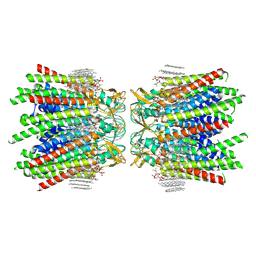 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN1
 
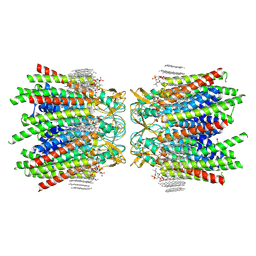 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
4DH2
 
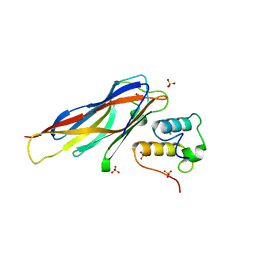 | | Crystal structure of Coh-OlpC(Cthe_0452)-Doc435(Cthe_0435) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin type 1, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
5VFC
 
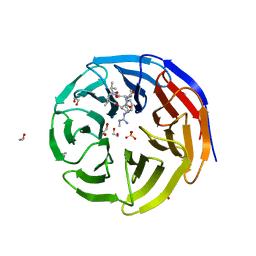 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
5VGB
 
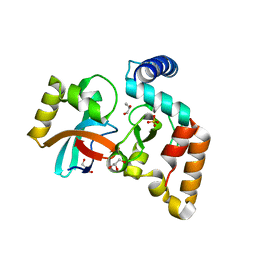 | | Crystal structure of NmeCas9 HNH domain bound to anti-CRISPR AcrIIC1 | | Descriptor: | Anti-CRISPR protein (AcrIIC1), CRISPR-associated endonuclease Cas9, GLYCEROL, ... | | Authors: | Harrington, L.B, Doxzen, K.W, Ma, E, Knott, G.J, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A Broad-Spectrum Inhibitor of CRISPR-Cas9.
Cell, 170, 2017
|
|
5VFX
 
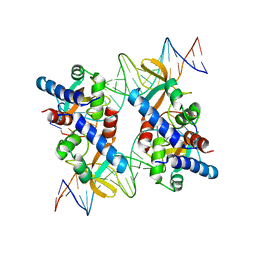 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
5VVL
 
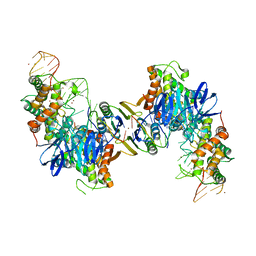 | | Cas1-Cas2 bound to full-site mimic with Ni | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (11-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5VZL
 
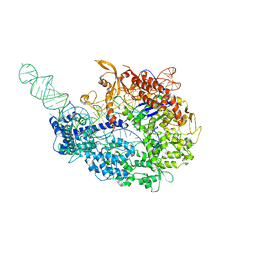 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
5W3V
 
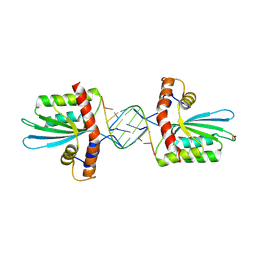 | | Crystal Structure of macaque APOBEC3H in complex with RNA | | Descriptor: | Apobec3H, RNA (5'-R(P*AP*AP*CP*CP*CP*CP*GP*GP*GP*C)-3'), RNA (5'-R(P*AP*AP*CP*CP*CP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Bohn, J.A, Thummar, K, York, A, Raymond, A, Brown, W.C, Bieniasz, P.D, Hatziioannou, T, Smith, J.L. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Nat Commun, 8, 2017
|
|
