6QV8
 
 | |
5FX2
 
 | |
3FX2
 
 | |
1VLX
 
 | | STRUCTURE OF ELECTRON TRANSFER (COBALT-PROTEIN) | | Descriptor: | AZURIN, COBALT (II) ION | | Authors: | Bonander, N, Vanngard, T, Tsai, L.-C, Langer, V, Nar, H, Sjolin, L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The metal site of Pseudomonas aeruginosa azurin, revealed by a crystal structure determination of the Co(II) derivative and Co-EPR spectroscopy.
Proteins, 27, 1997
|
|
1W84
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(2-PYRIDIN-4-YLETHYL)-1H-INDOLE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation.
J. Med. Chem., 48, 2005
|
|
1W83
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-CHLORO-3-(PYRIDIN-3-YLOXYMETHYL)-PHENYL]-3-FLUORO- | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation.
J. Med. Chem., 48, 2005
|
|
8E20
 
 | |
8EDM
 
 | |
7EST
 
 | |
3HE6
 
 | |
6DNA
 
 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
3HE7
 
 | | Crystal structure of mouse CD1d-alpha-galactosylceramide with mouse Valpha14-Vbeta7 NKT TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors
Immunity, 31, 2009
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
4FX2
 
 | |
4O2G
 
 | | Crystal structure of carbomonoxy murine neuroglobin mutant V140W | | Descriptor: | CARBON MONOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-17 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NZI
 
 | | Crystal structure of murine neuroglobin mutant V140W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-12 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O35
 
 | | Crystal structure of carbomonoxy murine neuroglobin mutant F106W | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CARBON MONOXIDE, Neuroglobin, ... | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2G1Q
 
 | | crystal structure of KSP in complex with inhibitor 9h | | Descriptor: | (5S)-5-(3-AMINOPROPYL)-3-(2,5-DIFLUOROPHENYL)-N-ETHYL-5-PHENYL-4,5-DIHYDRO-1H-PYRAZOLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-02-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 4: Structure-based design of 5-alkylamino-3,5-diaryl-4,5-dihydropyrazoles as potent, water-soluble inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4FQ0
 
 | |
6ZHY
 
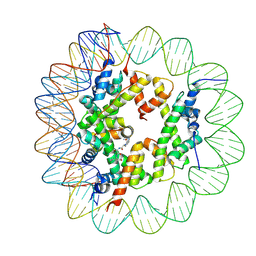 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
2GHL
 
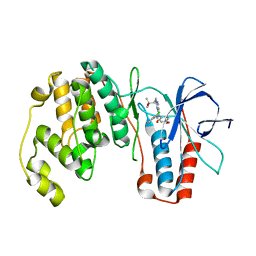 | | Mutant Mus Musculus P38 Kinase Domain in Complex with Inhibitor PG-874743 | | Descriptor: | 3-(2-CHLOROPHENYL)-1-(2-{[(1S)-2-HYDROXY-1,2-DIMETHYLPROPYL]AMINO}PYRIMIDIN-4-YL)-1-(4-METHOXYPHENYL)UREA, Mitogen-activated protein kinase 14 | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Brugel, T.A. | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Development of N-2,4-pyrimidine-N-phenyl-N'-phenyl ureas as inhibitors of tumor necrosis factor alpha (TNF-alpha) synthesis. Part 1.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5W1V
 
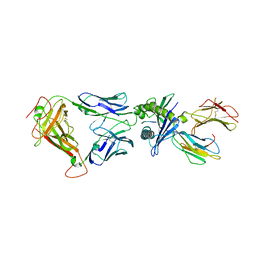 | | Structure of the HLA-E-VMAPRTLIL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
5W10
 
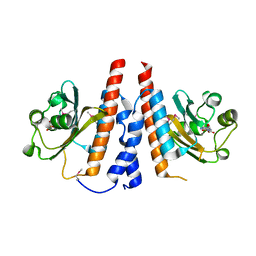 | | Lcd1 GAF domain in complex with cAMP ligand | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cGMP-specific phosphodiesterase | | Authors: | Guzzo, C.R, Maciel, N.K, Barbosa, A.S, Farah, C.S. | | Deposit date: | 2017-06-01 | | Release date: | 2017-06-28 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Enzymatic Characterization of a cAMP-Dependent Diguanylate Cyclase from Pathogenic Leptospira Species.
J. Mol. Biol., 429, 2017
|
|
6EST
 
 | |
2XTI
 
 | | Asparaginyl-tRNA synthetase from Brugia malayi complexed with ATP:Mg and L-Asp-beta-NOH adenylate:PPi:Mg | | Descriptor: | 5'-O-[(R)-{[(2S)-2-amino-4-(hydroxyamino)-4-oxobutanoyl]oxy}(hydroxy)phosphoryl]adenosine, ADENOSINE-5'-TRIPHOSPHATE, ASPARAGINYL-TRNA SYNTHETASE, ... | | Authors: | Crepin, T, Haertlein, M, Kron, M, Cusack, S. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Hybrid Structural Model of the Complete Brugia Malayi Cytoplasmic Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 405, 2011
|
|
