8RFK
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - single mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, Quinoprotein glucose dehydrogenase B | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RE0
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
6ELC
 
 | | Crystal Structure of O-linked Glycosylated VSG3 | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stebbins, C.E. | | Deposit date: | 2017-09-28 | | Release date: | 2018-07-11 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat.
Nat Microbiol, 3, 2018
|
|
6D5F
 
 | | Cryo-EM reconstruction of membrane-enveloped filamentous virus SFV1 (Sulfolobus filamentous virus 1) | | Descriptor: | DNA (336-MER), Fimbrial protein | | Authors: | Wang, F, Osinski, T, Liu, Y, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-08-29 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile.
Nat Commun, 9, 2018
|
|
2Q3X
 
 | | The RIM1alpha C2B domain | | Descriptor: | CHLORIDE ION, Regulating synaptic membrane exocytosis protein 1, SODIUM ION, ... | | Authors: | Guan, R, Dai, H, Tomchick, D.R, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2007-05-30 | | Release date: | 2007-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the RIM1alpha C(2)B Domain at 1.7 A Resolution.
Biochemistry, 46, 2007
|
|
5UG7
 
 | | Calcium bound Perforin C2 Domain - T431D | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5W1B
 
 | | Tryptophan indole-lyase | | Descriptor: | POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5DU1
 
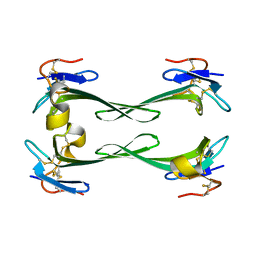 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P21 space group. | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
4ZIZ
 
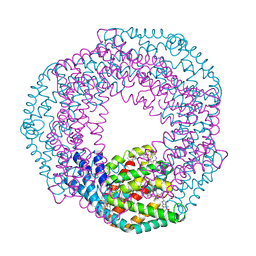 | | Serial Femtosecond Crystallography of Soluble Proteins in Lipidic Cubic Phase (C-Phycocyanin from T. elongatus) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Fromme, R, Ishchenko, A, Metz, M, Roy-Chowdhury, S, Basu, S, Boutet, S, Fromme, P, White, T.A, Barty, A, Spence, J.C.H, Weierstall, U, Liu, W, Cherezov, V. | | Deposit date: | 2015-04-28 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial femtosecond crystallography of soluble proteins in lipidic cubic phase.
IUCrJ, 2, 2015
|
|
5DZ5
 
 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P41212 space group | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
5IML
 
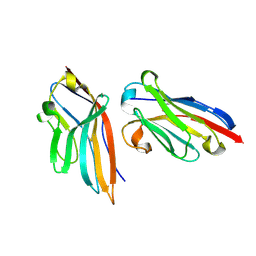 | | Nanobody targeting human Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IMK
 
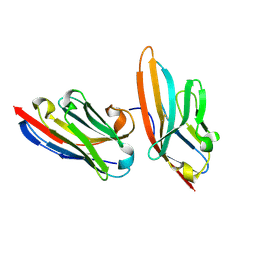 | | Nanobody targeting human Vsig4 in Spacegroup C2 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.227 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IMO
 
 | | Nanobody targeting mouse Vsig4 in Spacegroup P3221 | | Descriptor: | Nanobody, Protein Vsig4 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IMM
 
 | | Nanobody targeting mouse Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, Protein Vsig4 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
3EYW
 
 | | Crystal structure of the C-terminal domain of E. coli KefC in complex with KefF | | Descriptor: | C-terminal domain of Glutathione-regulated potassium-efflux system protein kefC fused to full length Glutathione-regulated potassium-efflux system ancillary protein kefF, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Roosild, T.P. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | KTN (RCK) Domains Regulate K(+) Channels and Transporters by Controlling the Dimer-Hinge Conformation.
Structure, 17, 2009
|
|
6T8P
 
 | | HKATII IN COMPLEX WITH LIGAND (2R)-N-benzyl-1-[6-methyl-5-(oxan-4-yl)-7-oxo-6H,7H-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(fluoranyl)-~{N}-[5-[(2~{R})-2-(3-fluorophenyl)-3-methyl-butyl]-1,3,4-thiadiazol-2-yl]benzenesulfonamide, IODIDE ION, ... | | Authors: | Blaesse, M, Venalainen, J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of sulfonamides and 9-oxo-2,8-diazaspiro[5,5]undecane-2-carboxamides as human kynurenine aminotransferase 2 (KAT2) inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2LC2
 
 | | Solution structure of the RXLR effector P. capsici AVR3a4 | | Descriptor: | AVR3a4 | | Authors: | Li, H, Koshiba, S, Yaeno, T, Sato, M, Watanabe, S, Harada, T, Shirasu, K, Kigawa, T. | | Deposit date: | 2011-04-12 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A PIP-binding interface in the oomycete RXLR effector AVR3A is required for its accumulation in host cells to modulate plant immunity
To be Published
|
|
6T8Q
 
 | | HKATII IN COMPLEX WITH LIGAND (2R)-N-benzyl-1-[6-methyl-5-(oxan-4-yl)-7-oxo-6H,7H-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]pyrrolidine-2-carboxamide | | Descriptor: | (2~{R})-1-[6-methyl-5-(oxan-4-yl)-7-oxidanylidene-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]-~{N}-(phenylmethyl)pyrrolidine-2-carboxamide, ACETATE ION, CADMIUM ION, ... | | Authors: | Blaesse, M, Venalainen, J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of sulfonamides and 9-oxo-2,8-diazaspiro[5,5]undecane-2-carboxamides as human kynurenine aminotransferase 2 (KAT2) inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7QQK
 
 | | TIR-SAVED effector bound to cA3 | | Descriptor: | RNA (5'-R(P*AP*AP*A)-3'), TIR_SAVED fusion protein | | Authors: | Spagnolo, L, White, M.F, Hogrel, G, Guild, A. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic nucleotide-induced helical structure activates a TIR immune effector.
Nature, 608, 2022
|
|
7V3W
 
 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, VpsR | | Authors: | Chakrabortty, T, Sen, U, Chowdhury, S.R. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
7V2V
 
 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | SULFATE ION, VpsR | | Authors: | Chakrabortty, T, Sen, U, Chowdhury, S.R. | | Deposit date: | 2021-08-10 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
6R1L
 
 | |
6FNE
 
 | | Structure of human Brag2 (Sec7-PH domains) with the inhibitor Bragsin bound to the PH domain | | Descriptor: | (2~{S})-6-methyl-5-nitro-2-(trifluoromethyl)-2,3-dihydrochromen-4-one, IQ motif and SEC7 domain-containing protein 1, NONAETHYLENE GLYCOL | | Authors: | Nawrotek, A, Zeghouf, M, Cherfils, J. | | Deposit date: | 2018-02-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | PH-domain-binding inhibitors of nucleotide exchange factor BRAG2 disrupt Arf GTPase signaling.
Nat.Chem.Biol., 15, 2019
|
|
