8JLJ
 
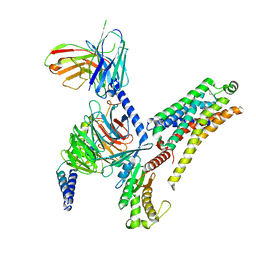 | | T1AM-bound mTAAR1-Gs protein complex | | Descriptor: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSO
 
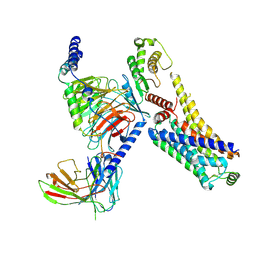 | | AMPH-bound hTAAR1-Gs protein complex | | Descriptor: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JEL
 
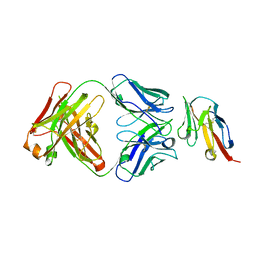 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form I | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEO
 
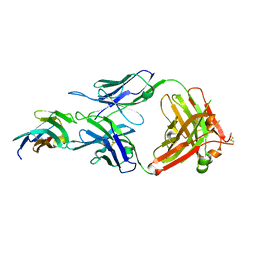 | | Crystal structure of TIGIT in complexed with Tiragolumab | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEN
 
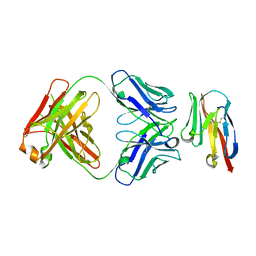 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form II | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEP
 
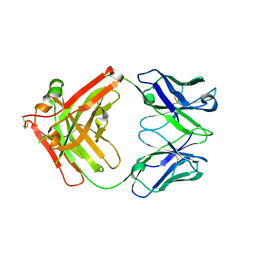 | | Crystal structure of Ociperlimab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEQ
 
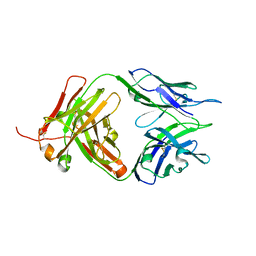 | | Crystal structure of Tiragolumab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
7T9U
 
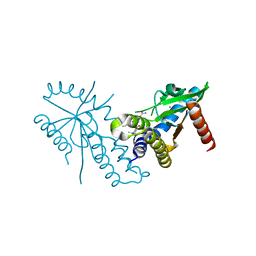 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
2GC4
 
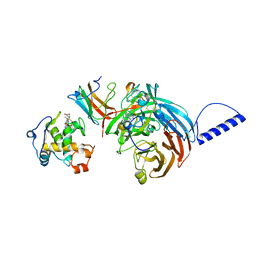 | | Structural comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution. | | Descriptor: | Amicyanin, COPPER (II) ION, Cytochrome c-L, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
2GC7
 
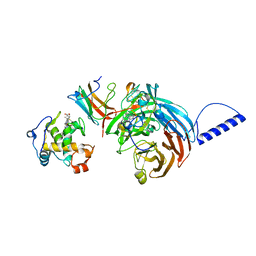 | | Substrate reduced, copper free complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans. | | Descriptor: | Amicyanin, Cytochrome c-L, HEME C, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structral comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
2I0X
 
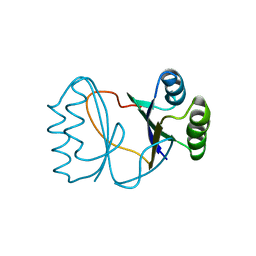 | | Hypothetical protein PF1117 from Pyrococcus furiosus | | Descriptor: | Hypothetical protein PF1117 | | Authors: | Chen, L.Q, Fu, Z.-Q, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein Pf1117 from Pyrococcus furiosus
To be Published
|
|
8JOV
 
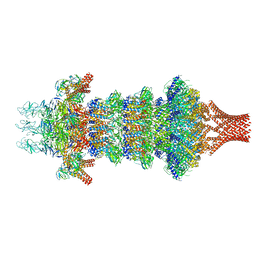 | | Portal-tail complex of phage GP4 | | Descriptor: | Portal protein, Putative tail fiber protein, Virion associated protein, ... | | Authors: | Liu, H, Chen, W. | | Deposit date: | 2023-06-08 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
4YDK
 
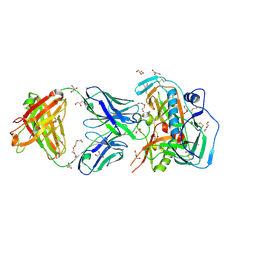 | | Crystal structure of broadly and potently neutralizing antibody C38-VRC16.01 in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4YDJ
 
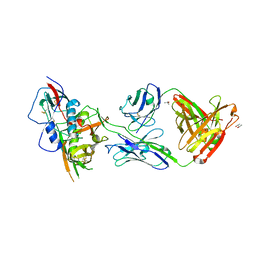 | | Crystal structure of broadly and potently neutralizing antibody 44-VRC13.01 in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4YDL
 
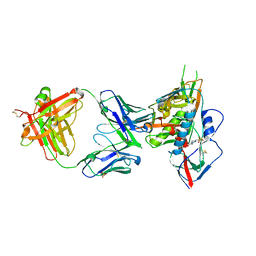 | | Crystal structure of broadly and potently neutralizing antibody C38-VRC18.02 in complex with HIV-1 clade AE strain 93TH057gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
4YDI
 
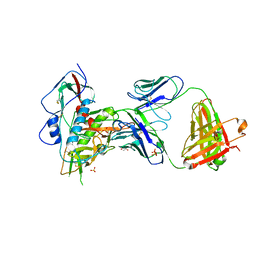 | | Crystal structure of broad and potently neutralizing VRC01-class antibody Z258-VRC27.01, isolated from human donor Z258, in complex with HIV-1 gp120 from clade A strain Q23.17 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.452 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
8I1V
 
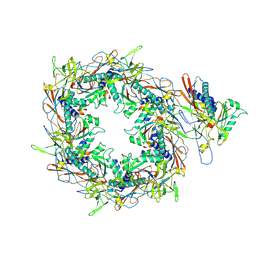 | | The asymmetric unit of P22 procapsid | | Descriptor: | Major capsid protein, Scaffolding protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8I1T
 
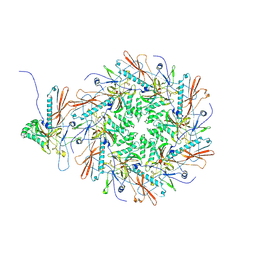 | | The asymmetric unit of P22 empty capsid | | Descriptor: | Major capsid protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8JOU
 
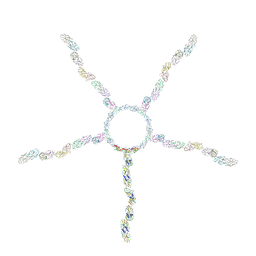 | | Fiber I and fiber-tail-adaptor of phage GP4 | | Descriptor: | Virion-associated phage protein, rope protein of phage GP4 | | Authors: | Liu, H, Chen, W. | | Deposit date: | 2023-06-08 | | Release date: | 2024-01-03 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
7EY7
 
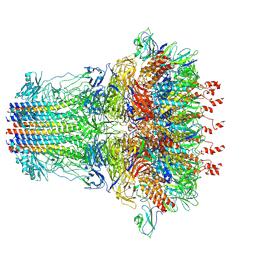 | | bacteriophage T7 tail complex | | Descriptor: | Internal virion protein gp14, Tail fiber protein, Tail tubular protein gp11, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY8
 
 | | portal | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY9
 
 | | tail proteins | | Descriptor: | Tail fiber protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY6
 
 | | The portal protein (GP8) of bacteriophage T7 | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
