1IG8
 
 | | Crystal Structure of Yeast Hexokinase PII with the correct amino acid sequence | | Descriptor: | SULFATE ION, hexokinase PII | | Authors: | Kuser, P.R, Krauchenco, S, Antunes, O.A, Polikarpov, I. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action.
J.Biol.Chem., 275, 2000
|
|
5T62
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5V2N
 
 | | Crystal Structure of APO Human SETD8 | | Descriptor: | 1,2-ETHANEDIOL, N-lysine methyltransferase KMT5A | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-05 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
5CZJ
 
 | |
4IM7
 
 | | Crystal structure of fructuronate reductase (ydfI) from E. coli CFT073 (EFI TARGET EFI-506389) complexed with NADH and D-mannonate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-MANNONIC ACID, Hypothetical oxidoreductase ydfI, ... | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2013-01-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of fructuronate reductase (ydfI) from E. coli CFT073 (EFI TARGET EFI-506389) complexed with NADH and D-mannonate
To be Published
|
|
4IL2
 
 | | Crystal structure of D-mannonate dehydratase (rspA) from E. coli CFT073 (EFI TARGET EFI-501585) | | Descriptor: | MAGNESIUM ION, Starvation sensing protein rspA | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-28 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mannonate degradation pathway in E. coli CFT073
To be Published
|
|
4ILK
 
 | | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MANGANESE (II) ION, Starvation sensing protein rspB, ... | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-31 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH
To be Published
|
|
4QVI
 
 | | Crystal structure of mutant ribosomal protein M218L TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 50S ribosomal protein L1, ACETATE ION, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6XDF
 
 | | Crystal structure of IRE1a in complex with G-4100 | | Descriptor: | 4-amino-N-(6-chloro-2-fluoro-3-{[(pyrrolidin-1-yl)sulfonyl]amino}phenyl)quinazoline-8-carboxamide, SODIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Steinbacher, S, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, M06 | | Authors: | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | Deposit date: | 2014-05-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
8K8A
 
 | | Crystal structure of NFIL3 in complex with TTACGTAA DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*G)-3'), Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K89
 
 | | Crystal structure of NFIL3 | | Descriptor: | Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K8D
 
 | |
8K86
 
 | | Crystal structure of NFIL3 in complex with TTATGTAA DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*TP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*AP*CP*AP*TP*AP*AP*TP*G)-3'), Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
8K8C
 
 | | Crystal structure of C/EBPalpha BZIP domain bound to a high affinity DNA | | Descriptor: | CCAAT/enhancer-binding protein alpha, DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*T)-3'), ... | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
7MWI
 
 | | Crystal structure of human BAZ2A | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
8K3L
 
 | | SOD1 and Nanobody3 complex | | Descriptor: | COPPER (II) ION, NB3, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cheng, S, Liu, R, Ding, Y. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SOD1 and Nanobody1 complex
To Be Published
|
|
6BOZ
 
 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
6PI7
 
 | |
6ZUE
 
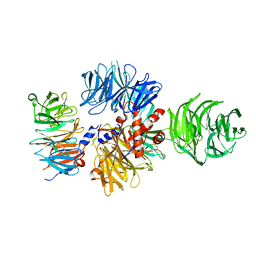 | |
5VFI
 
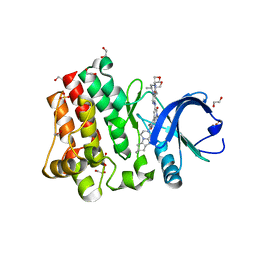 | | Bruton's tyrosine kinase (BTK) with GDC-0853 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3'-(hydroxymethyl)-1-methyl-5-({5-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-2-yl}amino)-6-oxo[1,6-dihydro[3,4'-bipyridine]]-2'-yl]-7,7-dimethyl-3,4,7,8-tetrahydro-2H-cyclopenta[4,5]pyrrolo[1,2-a]pyrazin-1(6H)-one, SULFATE ION, ... | | Authors: | Steinbacher, S, Eigenbrot, C. | | Deposit date: | 2017-04-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of GDC-0853: A Potent, Selective, and Noncovalent Bruton's Tyrosine Kinase Inhibitor in Early Clinical Development.
J. Med. Chem., 61, 2018
|
|
5HZB
 
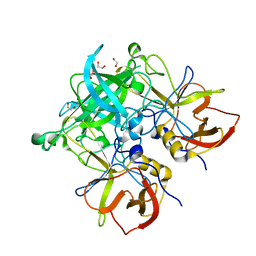 | | Crystal structure of GII.10 P domain in complex with 2-fucosyllactose (2'FL) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hansman, G.S, Koromyslova, A.D, Singh, B.K.S. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides.
J.Virol., 90, 2016
|
|
5HZA
 
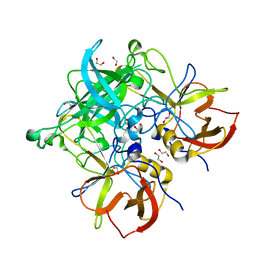 | | Crystal structure of GII.10 P domain in complex with 3-fucosyllactose (3 FL) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]beta-D-glucopyranose | | Authors: | Hansman, G.S, Koromyslova, A.D, Singh, B.K. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides.
J.Virol., 90, 2016
|
|
7LFV
 
 | |
