6VNS
 
 | | Crystal structure of TYK2 kinase with compound 13 | | Descriptor: | (1R,2R)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNY
 
 | | Crystal structure of TYK2 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
3M6F
 
 | | CD11A I-domain complexed with 6-((5S,9R)-9-(4-CYANOPHENYL)-3-(3,5-DICHLOROPHENYL)-1-METHYL-2,4-DIOXO-1,3,7- TRIAZASPIRO[4.4]NON-7-YL)NICOTINIC ACID | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, NITRATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small molecule antagonist of leukocyte function associated antigen-1 (LFA-1): structure-activity relationships leading to the identification of 6-((5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]nonan-7-yl)nicotinic acid (BMS-688521).
J.Med.Chem., 53, 2010
|
|
6VNX
 
 | | Crystal structure of TYK2 kinase with compound 19 | | Descriptor: | (1S)-2,2-difluoro-N-[(1S,5R,6R)-3-{5-fluoro-2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-6-methyl-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6W8L
 
 | | Crystal structure of JAK1 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNV
 
 | | Crystal structure of TYK2 kinase with compound 14 | | Descriptor: | (1S,2S)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
7F3B
 
 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
6OAP
 
 | | Crystal structure of a dual sensor histidine kinase in the green-light absorbing Pg state | | Descriptor: | Dual sensor histidine kinase, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OAQ
 
 | | Crystal structure of a dual sensor histidine kinase in BeF3- bound state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Dual sensor histidine kinase, MAGNESIUM ION, ... | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OB8
 
 | | Crystal structure of a dual sensor histidine kinase in green light illuminated state | | Descriptor: | Dual sensor histidine kinase, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5BJS
 
 | | Apo ctPRC2 in an autoinhibited state | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein SUZ12, Polycomb Protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2016-10-22 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
6U3U
 
 | | Crystal Structure of Shiga Toxin 2K | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Shiga toxin 2K subunit A, ... | | Authors: | Zhang, Y.Z, He, X.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2.
Microorganisms, 8, 2019
|
|
6OZB
 
 | | Crystal structure of the phycoerythrobilin-bound GAF domain from a cyanobacterial phytochrome | | Descriptor: | PHYCOERYTHROBILIN, Two-component sensor histidine kinase | | Authors: | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OZA
 
 | | Crystal structure of the phycocyanobilin-bound GAF domain from a cyanobacterial phytochrome | | Descriptor: | PHYCOCYANOBILIN, Two-component sensor histidine kinase | | Authors: | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TQR
 
 | |
7BW6
 
 | | Varicella-zoster virus capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Wang, P.Y, Qi, J.X, Liu, C.C, Sun, J.Q. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the varicella-zoster virus A-capsid.
Nat Commun, 11, 2020
|
|
7T7A
 
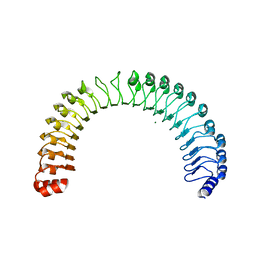 | | Crystal Structure of Human SHOC2: A Leucine-Rich Repeat Protein | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, NITRATE ION | | Authors: | Hajian, B, Lemke, C, Kwon, J, Bian, Y, Fuller, C, Aguirre, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex.
Nature, 609, 2022
|
|
6U4Y
 
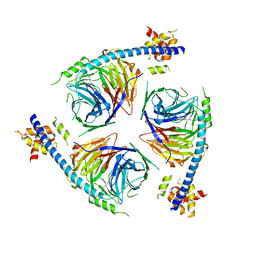 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5BPE
 
 | | Crystal structure of EV71 3Cpro in complex with a potent and selective Inhibitor | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{S})-1-cyano-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-3-phenyl-2-[[(~{E})-3-phenylprop-2-enoyl]amino]propanamide, EV71 3Cpro | | Authors: | Luqing, S, Yin, Z. | | Deposit date: | 2015-05-28 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanohydrin as an Anchoring Group for Potent and Selective Inhibitors of Enterovirus 71 3C Protease
J.Med.Chem., 58, 2015
|
|
5UBR
 
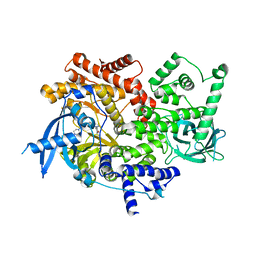 | | CRYSTAL STRUCTURE OF PI3K ALPHA IN COMPLEX WITH A 7-(3-(PIPERAZIN-1-YL)PHENYL)PYRROLO[2,1-F][1,2,4] TRIAZIN-4-AMINE DERIVIATINE | | Descriptor: | 1-[4-(3-{4-amino-5-[1-(oxan-4-yl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)piperazin-1-yl]ethan-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Sack, J.S. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of 7-(3-(piperazin-1-yl)phenyl)pyrrolo[2,1-f][1,2,4]triazin-4-amine derivatives as highly potent and selective PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4QQG
 
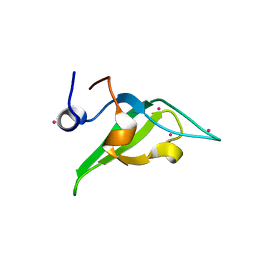 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
5UBT
 
 | | CRYSTAL STRUCTURE OF PI3K DELTA IN COMPLEX WITH A 7-(3-(PIPERAZIN-1-YL)PHENYL)PYRROLO[2,1-F][1,2,4] TRIAZIN-4-AMINE DERIVIATINE | | Descriptor: | 1-[4-(3-{4-amino-5-[1-(oxan-4-yl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)piperazin-1-yl]ethan-1-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Sack, J.S. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of 7-(3-(piperazin-1-yl)phenyl)pyrrolo[2,1-f][1,2,4]triazin-4-amine derivatives as highly potent and selective PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UF7
 
 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
7SHP
 
 | | Crystal structure of hSTING in complex with c[2',3'-(ribo-2'-G, xylo-3'-A)-MP](RJ244) | | Descriptor: | (2S,5R,7R,8R,10S,12aR,14R,15R,15aR,16R)-7-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
