8HND
 
 | | Cryo-EM structure of human OATP1B1 in complex with estrone-3-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
4WE8
 
 | | The crystal structure of hemagglutinin of influenza virus A/Victoria/361/2011 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
8AMH
 
 | |
4WDR
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WE5
 
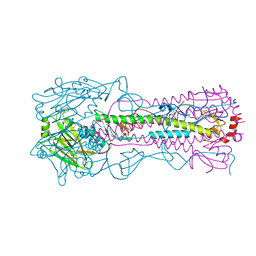 | | The crystal structure of hemagglutinin from A/Port Chalmers/1/1973 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
8HL5
 
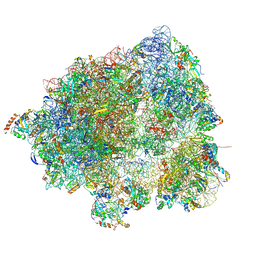 | |
8AN6
 
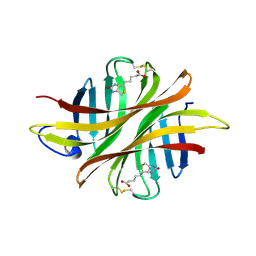 | |
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1I
 
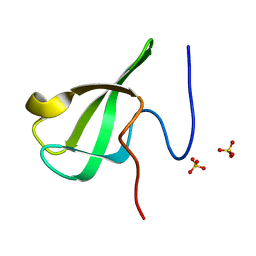 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
4V83
 
 | | Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4CND
 
 | | Crystal structure of E.coli TrmJ | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (CYTIDINE/URIDINE-2'-O-)-METHYLTRANSFERASE TRMJ | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
7NAE
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
4UYB
 
 | | Crystal structure of SEC14-like protein 3 | | Descriptor: | 1,2-ETHANEDIOL, SEC14-LIKE PROTEIN 3, UNKNOWN LIGAND | | Authors: | Kopec, J, Goubin, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Sec14-Like Protein 3
To be Published
|
|
7MME
 
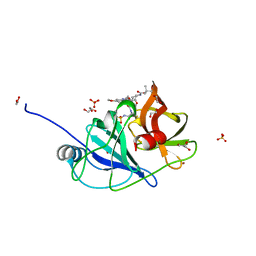 | |
6L97
 
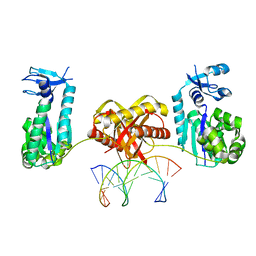 | | Complex of DNA polymerase IV and L-DNA duplex | | Descriptor: | DNA (5'-D(*(0DG)P*(0DG)P*(0DG)P*(0DG)P*(0DG)P*(0DA)P*(0DA)P*(0DG)P*(0DG)P*(0DA)P*(0DT)P*(0DT)P*(0DC)P*(0DC))-3'), DNA (5'-D(P*(0DG)P*(0DG)P*(0DA)P*(0DA)P*(0DT)P*(0DC)P*(0DC)P*(0DT)P*(0DT)P*(0DC)P*(0DC)P*(0DC)P*(0DC)P*(0DC))-3'), DNA polymerase IV | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
4URG
 
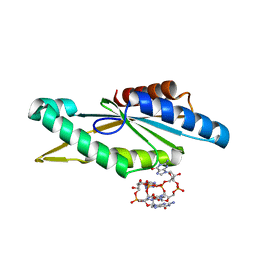 | | Crystal Structure of GGDEF domain from T.maritima (active-like dimer) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
2VDN
 
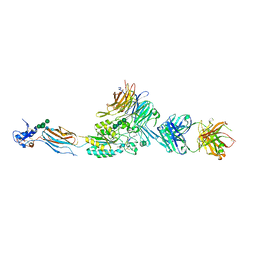 | | Re-refinement of Integrin AlphaIIbBeta3 Headpiece Bound to Antagonist Eptifibatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EPTIFIBATIDE, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
7N3Y
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant in Complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Wojtaszek, J.L, Krahn, J, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | Descriptor: | Glycoprotein,Glycoprotein,Glycoprotein | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
7MST
 
 | | Phosphorylated human E105Qa GTP-specific succinyl-CoA synthetase complexed with coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2021-05-12 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structure of succinyl-CoA synthetase bound to the succinyl-phosphate intermediate clarifies the catalytic mechanism of ATP-citrate lyase
Acta Crystallogr.,Sect.F, 78, 2022
|
|
4V1U
 
 | |
5XVC
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
4W5N
 
 | | The Crystal Structure of Human Argonaute2 Bound to a Defined Guide RNA | | Descriptor: | MAGNESIUM ION, PHENOL, Protein argonaute-2, ... | | Authors: | Schirle, N.T, Sheu-Gruttadauria, J, MacRae, I.J. | | Deposit date: | 2014-08-18 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gene regulation. Structural basis for microRNA targeting.
Science, 346, 2014
|
|
4V2S
 
 | | Crystal structure of Hfq in complex with the sRNA RydC | | Descriptor: | RNA-BINDING PROTEIN HFQ, RYDC | | Authors: | Dimastrogiovanni, D, Frohlich, K.S, Bruce, H.A, Bandyra, K.J, Hohensee, S, Vogel, J, Luisi, B.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Recognition of the small regulatory RNA RydC by the bacterial Hfq protein.
Elife, 3, 2014
|
|
4W5X
 
 | |
