5AER
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with D2 dopamine receptor peptide from Homo sapiens | | Descriptor: | CALCIUM ION, D(2) DOPAMINE RECEPTOR, NEURONAL CALCIUM SENSOR 1, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5AFP
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with rhodopsin kinase peptide from Homo sapiens | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, RHODOPSIN KINASE, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5AEQ
 
 | | Neuronal calcium sensor (NCS-1)from Rattus norvegicus | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, SODIUM ION | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
4JRR
 
 | | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Shumilin, I.A, Jameson-Lee, M, Cymborowski, M, Domagalski, M.J, Chertihin, O, Kpadeh, Z.Z, Yeh, A.J, Hoffman, P.S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila
To be Published
|
|
6Z3B
 
 | | Low resolution structure of RgNanOx | | Descriptor: | CITRIC ACID, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
2RGT
 
 | | Crystal Structure of Lhx3 LIM domains 1 and 2 with the binding domain of Isl1 | | Descriptor: | Fusion of LIM/homeobox protein Lhx3, linker, Insulin gene enhancer protein ISL-1, ... | | Authors: | Bhati, M, Lee, M, Guss, J.M, Matthews, J.M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes.
Embo J., 27, 2008
|
|
4Q6Q
 
 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6N
 
 | | Structural analysis of the tripeptide-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6M
 
 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6O
 
 | | Structural analysis of the mDAP-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6P
 
 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4RNZ
 
 | | Structure of Helicobacter pylori Csd3 from the hexagonal crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, NICKEL (II) ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RNY
 
 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3LZC
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | Descriptor: | Dph2 | | Authors: | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
3LZD
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
3MJM
 
 | | His257Ala mutant of dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Ernberg, K.E, Guss, J.M, Lee, M, Maher, M.J. | | Deposit date: | 2010-04-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | His257Ala mutant of dihydroorotase from E. coli
To be Published
|
|
1B0S
 
 | | BINDING OF AR-1-144, A TRI-IMIDAZOLE DNA MINOR GROOVE BINDER, TO CCGG SEQUENCE ANALYZED BY NMR SPECTROSCOPY | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, DNA (5'-D(*GP*AP*AP*CP*CP*GP*GP*TP*TP*C)-3') | | Authors: | Yang, X.-L, Kaenzig, C, Lee, M, Wang, A.H.-J. | | Deposit date: | 1998-11-12 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Binding of AR-1-144, a tri-imidazole DNA minor groove binder, to CCGG sequence analyzed by NMR spectroscopy.
Eur.J.Biochem., 263, 1999
|
|
1A50
 
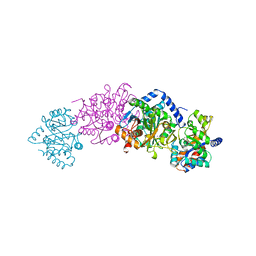 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH 5-FLUOROINDOLE PROPANOL PHOSPHATE | | Descriptor: | 5-FLUOROINDOLE PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
1A5S
 
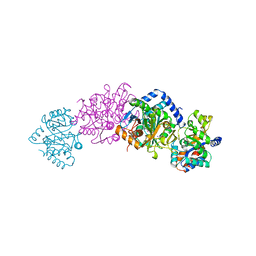 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH 5-FLUOROINDOLE PROPANOL PHOSPHATE AND L-SER BOUND AS AMINO ACRYLATE TO THE BETA SITE | | Descriptor: | 5-FLUOROINDOLE PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SERINE, ... | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-17 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
6UWZ
 
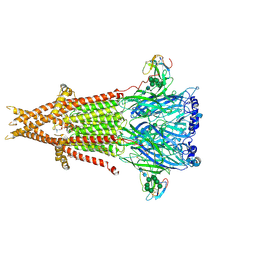 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with alpha-bungarotoxin | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Rahman, M.M, Teng, J, Worrell, B.T, Noveillo, C.M, Lee, M, Karlin, A, Stowell, M, Hibbs, R.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-04-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins.
Neuron, 106, 2020
|
|
1CYZ
 
 | | NMR STRUCTURE OF THE GAACTGGTTC/TRI-IMIDAZOLE POLYAMIDE COMPLEX | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, 5'-D(*GP*AP*AP*CP*TP*GP*GP*TP*TP*C)-3' | | Authors: | Yang, X.-L, Hubbard IV, R.B, Lee, M, Tao, Z.-F, Sugiyama, H, Wang, A.H.-J. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Imidazole-imidazole pair as a minor groove recognition motif for T:G mismatched base pairs
Nucleic Acids Res., 27, 1999
|
|
6UE0
 
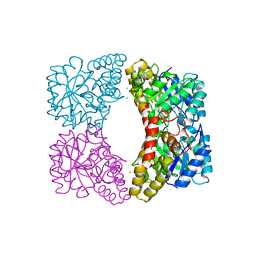 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
6DLN
 
 | |
6X44
 
 | | High Resolution Crystal Structure Analysis of SERA5 proenzyme from plasmodium falciparum | | Descriptor: | DI(HYDROXYETHYL)ETHER, Serine repeat antigen 5 | | Authors: | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | Deposit date: | 2020-05-22 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19733953 Å) | | Cite: | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
6X42
 
 | | High Resolution Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, ... | | Authors: | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
