8Q6S
 
 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Putative acetyl xylan esterase, ... | | Authors: | Mazurkewich, S, Seveso, A, Banerjee, S, Lo Leggio, L, Larsbrink, J. | | Deposit date: | 2023-08-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
2B5B
 
 | | A reptilian defensin with anti-bacterial and anti-viral activity | | Descriptor: | Defensin | | Authors: | Chattopadhyay, S, Sinha, N.K, Banerjee, S, Roy, D, Chattopadhyay, D, Roy, S. | | Deposit date: | 2005-09-28 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Small cationic protein from a marine turtle has beta-defensin-like fold and antibacterial and antiviral activity.
Proteins, 64, 2006
|
|
4FM5
 
 | | X-ray structure of des-methylflurbiprofen bound to murine COX-2 | | Descriptor: | (2-fluorobiphenyl-4-yl)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Xu, S, Banerjee, S, Windsor, M.A, Marnett, L.J. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Substrate-Selective Inhibition of Cyclooxygenase-2: Development and Evaluation of Achiral Profen Probes.
ACS Med Chem Lett, 3, 2012
|
|
7ZE9
 
 | | Structure of an AA16 LPMO-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Huang, Z, Banerjee, S, Muderspach, S.J, Sun, P, van Berkel, W.J.H, Kabel, M.A, Lo Leggio, L. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila.
Acs Catalysis, 13, 2023
|
|
7ZJ2
 
 | |
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JCZ
 
 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3J7H
 
 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JD2
 
 | | Glutamate dehydrogenase in complex with NADH, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
7MC6
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC5
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6N47
 
 | | The structure of SB-2-204-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloropyrido[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | Arnst, K, Banerjee, S, Wang, Y, Li, W, Miller, D, Li, W. | | Deposit date: | 2018-11-17 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray Crystal Structure Guided Discovery and Antitumor Efficacy of Dihydroquinoxalinone as Potent Tubulin Polymerization Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
4YJ0
 
 | | Crystal structure of the DM domain of human DMRT1 bound to 25mer target DNA | | Descriptor: | DNA (25-MER), Doublesex- and mab-3-related transcription factor 1, ZINC ION | | Authors: | Murphy, M.W, Lee, J.K, Rojo, S, Gearhart, M.D, Kurahashi, K, Banerjee, S, Loeuille, G, Bashamboo, A, McElreavey, K, Zarkower, D, Aihara, H, Bardwell, V.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | An ancient protein-DNA interaction underlying metazoan sex determination.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4Z0L
 
 | | The murine cyclooxygenase-2 complexed with a nido-dicarbaborate-containing indomethacin derivative | | Descriptor: | (R)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, (S)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Neumann, W, Banerjee, S, Hey-Hawkins, E, Marnett, L.J. | | Deposit date: | 2015-03-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | nido-Dicarbaborate Induces Potent and Selective Inhibition of Cyclooxygenase-2.
Chemmedchem, 11, 2016
|
|
4EBD
 
 | | Conformationally Restrained North-methanocarba-2'-deoxyadenosine Corrects the Error-Prone Nature of Human DNA Polymerase Iota | | Descriptor: | 5'-D(P*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(P*CP*TP*GP*GP*GP*TP*CP*CP*T)-3', CALCIUM ION, ... | | Authors: | Eoff, R.L, Ketkar, A, Banerjee, S. | | Deposit date: | 2012-03-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.571 Å) | | Cite: | A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota.
J.Am.Chem.Soc., 134, 2012
|
|
4EBE
 
 | | Conformationally Restrained North-methanocarba-2'-deoxyadenosine Corrects the Error-Prone Nature of Human DNA Polymerase Iota | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(P*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(P*CP*TP*GP*GP*GP*TP*CP*CP*T)-3', ... | | Authors: | Eoff, R.L, Ketkar, A, Banerjee, S. | | Deposit date: | 2012-03-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota.
J.Am.Chem.Soc., 134, 2012
|
|
2KZT
 
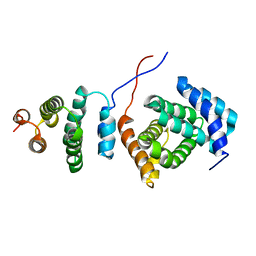 | | Structure of the Tandem MA-3 Region of Pdcd4 | | Descriptor: | Programmed cell death protein 4 | | Authors: | Waters, L.C, Strong, S.L, Oka, O, Muskett, F.W, Veverka, V, Banerjee, S, Schmedt, T, Henry, A.J, Klempnauer, K.H, Carr, M.D. | | Deposit date: | 2010-06-24 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the tandem MA-3 region of Pdcd4 protein and characterization of its interactions with eIF4A and eIF4G: molecular mechanisms of a tumor suppressor
J.Biol.Chem., 286, 2011
|
|
4EBC
 
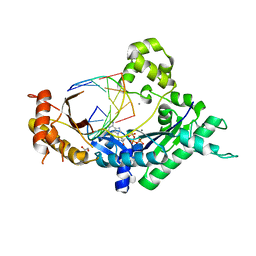 | | Conformationally Restrained North-methanocarba-2'-deoxyadenosine Corrects the Error-Prone Nature of Human DNA Polymerase Iota | | Descriptor: | 5'-D(P*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(P*CP*TP*GP*GP*GP*TP*CP*CP*T)-3', CALCIUM ION, ... | | Authors: | Eoff, R.L, Ketkar, A, Banerjee, S. | | Deposit date: | 2012-03-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota.
J.Am.Chem.Soc., 134, 2012
|
|
6P7A
 
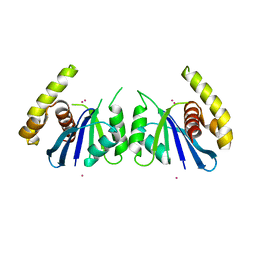 | | CRYSTAL STRUCTURE OF THE FOWLPOX VIRUS HOLLIDAY JUNCTION RESOLVASE | | Descriptor: | CADMIUM ION, Holliday junction resolvase | | Authors: | Li, N, Shi, K, Banerjee, S, Rao, T, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
6P7B
 
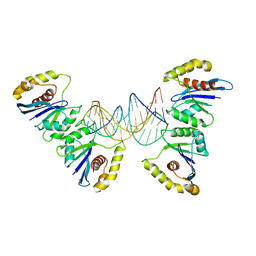 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | Descriptor: | DNA (29-MER), Holliday junction resolvase | | Authors: | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.317 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
6E5T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | Descriptor: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6E6M
 
 | | Crystal structure of human cellular retinol-binding protein 1 in complex with cannabidiorcin (CBDO) | | Descriptor: | (1'R,2'R)-4,5'-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,6-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
