4IS9
 
 | | Crystal Structure of the Escherichia coli LpxC/L-161,240 complex | | Descriptor: | (4R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydro-1,3-oxazole-4-carboxamide, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4J72
 
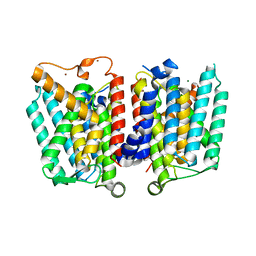 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
4LCH
 
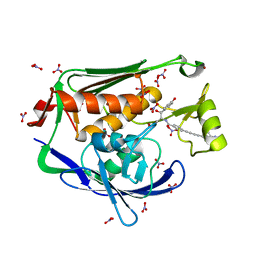 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | Descriptor: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LCF
 
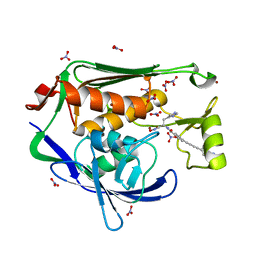 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-014 complex | | Descriptor: | NITRATE ION, Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-histidinamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LKV
 
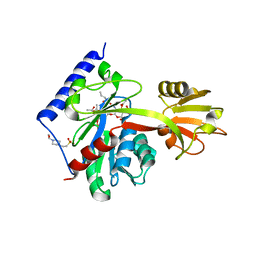 | | Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Emptage, R.P, Tonthat, N.K, York, J.D, Schumacher, M.A, Zhou, P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5109 Å) | | Cite: | Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
J.Biol.Chem., 289, 2014
|
|
2B5L
 
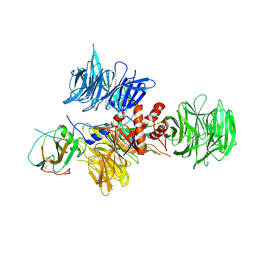 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | Descriptor: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2B5M
 
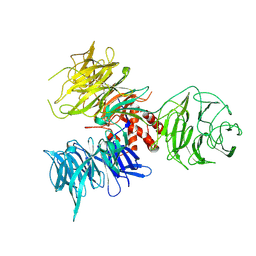 | | Crystal Structure of DDB1 | | Descriptor: | damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2B5N
 
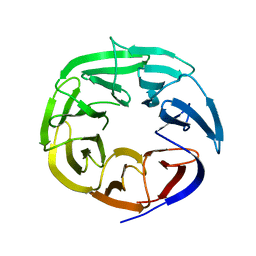 | | Crystal Structure of the DDB1 BPB Domain | | Descriptor: | ISOPROPYL ALCOHOL, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2LSJ
 
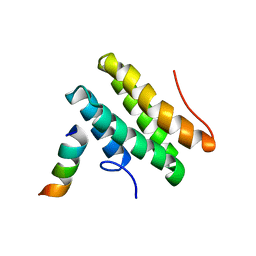 | |
2LSG
 
 | |
2MRM
 
 | |
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
4FQG
 
 | | Crystal structure of the TCERG1 FF4-6 tandem repeat domain | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Transcription elongation regulator 1 | | Authors: | Liu, J, Fan, S, Lee, C.J, Greenleaf, A.L, Zhou, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat.
J.Biol.Chem., 288, 2013
|
|
3P3E
 
 | | Crystal Structure of the PSEUDOMONAS AERUGINOSA LpxC/LPC-009 complex | | Descriptor: | N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, NITRATE ION, SODIUM ION, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
3P3G
 
 | | Crystal Structure of the Escherichia coli LpxC/LPC-009 complex | | Descriptor: | 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
3PS2
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-012 complex | | Descriptor: | 4-[4-(3-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3PS1
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-011 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3PS3
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-053 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3QC1
 
 | | Protein Phosphatase Subunit: Alpha4 | | Descriptor: | Immunoglobulin-binding protein 1 | | Authors: | Spiller, B.W, LeNoue-Newton, M.L, Watkins, G.R, Germane, K.L, Zhou, P, McCorvey, L.R, Wadzinski, B.E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Mid1 and PP2Ac binding domains of Alpha4 are both required for Alpha4 to inhibit PP2Ac degradation
TO BE PUBLISHED
|
|
3P3C
 
 | | Crystal Structure of the Aquifex aeolicus LpxC/LPC-009 complex | | Descriptor: | N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAD
 
 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAE
 
 | |
