5L04
 
 | |
3NS8
 
 | |
6F2N
 
 | | Crystal structure of BCII Metallo-beta-lactamase in complex with KDU197 | | Descriptor: | (~{Z})-3-[2-(naphthalen-2-ylmethyl)phenyl]-2-sulfanyl-prop-2-enoic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | McDonough, M.A, El-Hussein, A, Schofield, C.J, Zhang, D, Brem, J. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6EUM
 
 | | CRYSTAL STRUCTURE OF BCII METALLO-BETA-LACTAMASE IN COMPLEX WITH DZ-307 | | Descriptor: | (~{Z})-2-sulfanyl-3-[2,3,6-tris(fluoranyl)phenyl]prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6EWE
 
 | | Crystal structure of BCII Metallo-beta-lactamase in complex with DZ-308 | | Descriptor: | (~{Z})-3-(1-benzothiophen-3-yl)-2-sulfanyl-prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
2MRO
 
 | |
2MR9
 
 | |
8P3X
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Z
 
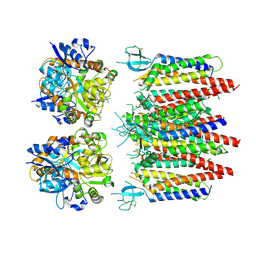 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Y
 
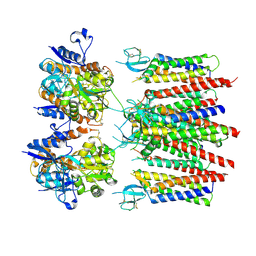 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8UOR
 
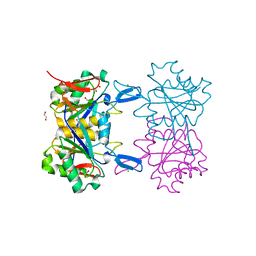 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19E) | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP9
 
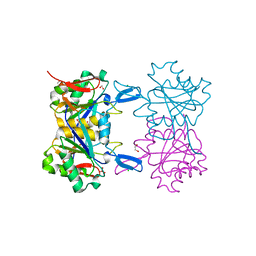 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19Q) | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, CHLORIDE ION, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UPC
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K158M) | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP6
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19A) in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, TETRAETHYLENE GLYCOL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOU
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOO
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP7
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19A) | | Descriptor: | Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP8
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F, complex with L-Asp) | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOW
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Asparaginase | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP3
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
6M24
 
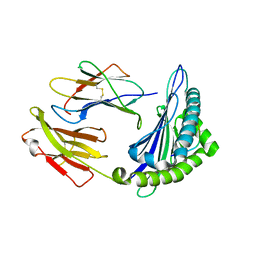 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-2 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2J
 
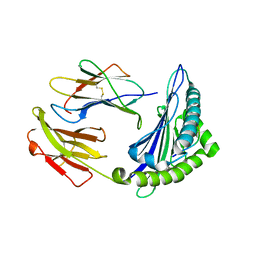 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-1 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2K
 
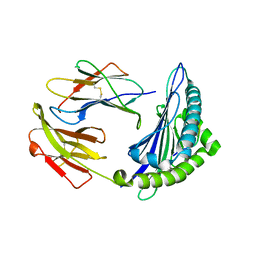 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-10 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
5OPC
 
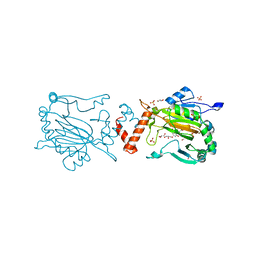 | | Factor Inhibiting HIF (FIH) in complex with zinc and Vadadustat | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Lu, X, Zhang, D. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5OX6
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with Vadadustat | | Descriptor: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Zhang, D, Schofield, C.J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
