7SQ7
 
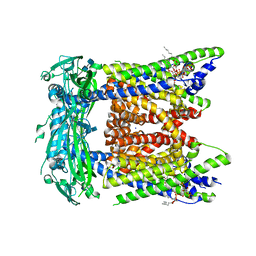 | | Cryo-EM structure of mouse PI(3,5)P2-bound TRPML1 channel at 2.41 Angstrom resolution | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ8
 
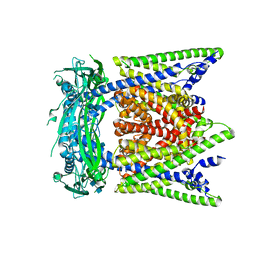 | | Cryo-EM structure of mouse apo TRPML1 channel at 2.598 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.598 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ9
 
 | | Cryo-EM structure of mouse temsirolimus/PI(3,5)P2-bound TRPML1 channel at 2.11 Angstrom resolution | | Descriptor: | (1R,2R,4S)-4-{(2R)-2-[(3S,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30S,34aS)-9,27-dihydroxy-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-1,5,11,28,29-pentaoxo-1,4,5,6,9,10,11,12,13,14,21,22,23,24,25,26,27,28,29,31,32,33,34,34a-tetracosahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontin-3-yl]propyl}-2-methoxycyclohexyl 3-hydroxy-2-(hydroxymethyl)-2-methylpropanoate, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ6
 
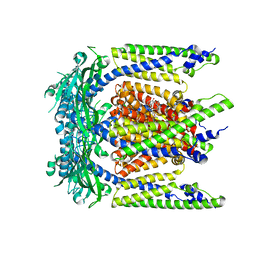 | | Cryo-EM structure of mouse agonist ML-SA1-bound TRPML1 channel at 2.32 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4O26
 
 | |
6VWC
 
 | | Crystal structure of Bcl-xL in complex with tetrahydroisoquinoline-pyridine based inhibitors | | Descriptor: | 6-{8-[(1,3-benzothiazol-2-yl)carbamoyl]-3,4-dihydroisoquinolin-2(1H)-yl}-3-{1-[(pyridin-4-yl)methyl]-1H-pyrazol-4-yl}pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Judge, R.A, Judd, A.S. | | Deposit date: | 2020-02-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Discovery of A-1331852, a First-in-Class, Potent, and Orally-Bioavailable BCL-X L Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
7EWA
 
 | | GDP-bound KRAS G12D in complex with TH-Z827 | | Descriptor: | 4-[(1~{R},5~{S})-3,8-diazabicyclo[3.2.1]octan-8-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
5VKC
 
 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
6E66
 
 | |
4QVX
 
 | |
9IJ1
 
 | | Cryo-EM Structure of MILI-piRNA-target (22-nt, bilobed) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (26-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IIY
 
 | | Cryo-EM Structure of EfPiwi-piRNA-target (25-nt, bilobed) | | Descriptor: | Piwi, RNA (5'-R(P*CP*GP*UP*CP*UP*AP*UP*AP*CP*AP*AP*CP*CP*GP*AP*UP*CP*AP*GP*CP*U)-3'), RNA (5'-R(P*UP*AP*GP*CP*AP*GP*AP*UP*CP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*AP*CP*G)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ4
 
 | | Cryo-EM Structure of MILI(K852A)-piRNA-target (26-nt) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ5
 
 | | Cryo-EM Structure of MILI(K853A)-piRNA-target | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ3
 
 | | Cryo-EM Structure of MILI-piRNA-target (26-nt) | | Descriptor: | MANGANESE (II) ION, Piwi-like protein 2, RNA (25-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ0
 
 | | Cryo-EM Structure of MILI-piRNA-target (8-nt) | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (26-MER), ... | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IJ2
 
 | | Cryo-EM Structure of MILI-piRNA-target (22-nt, comma) | | Descriptor: | Piwi-like protein 2, RNA (5'-R(P*CP*AP*AP*GP*UP*UP*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*A)-3'), RNA (5'-R(P*UP*UP*AP*CP*CP*AP*UP*CP*AP*AP*CP*AP*UP*GP*GP*AP*AP*AP*CP*UP*UP*G)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
9IIZ
 
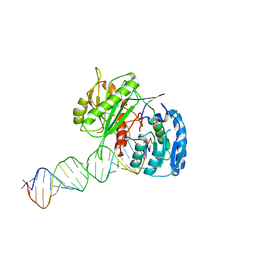 | | Cryo-EM Structure of EfPiwi-piRNA-target (25-nt, comma) | | Descriptor: | Piwi, RNA (25-MER), RNA (5'-R(P*AP*GP*CP*CP*AP*AP*GP*UP*UP*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*A)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
7YG6
 
 | | Cryo-EM structure of the EfPiwi(N959K) in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFX
 
 | | Cryo-EM structure of Hili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YGN
 
 | | Cryo-EM structure of the Mili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFQ
 
 | | Cryo-EM structure of the EfPiwi (N959K)-piRNA-target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi, RNA (5'-R(*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFY
 
 | | Cryo-EM structure of the Mili-piRNA- target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (5'-R(P*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
8WEC
 
 | | Crystal structure of Arabidopsis thaliana MIK2 ectodomain in complex with BAK1 ectodomain and SCOOP12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation.
Nat.Plants, 10, 2024
|
|
