7DB9
 
 | | IC1 in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(2,4,6-TRIMETHOXY-PHENYL)-METHYLENE]-INDOLIN-2-ONE, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | IC1 in complex with tubulin
To Be Published
|
|
7DBA
 
 | | RYX in complex with tubulin | | Descriptor: | 2-(1-methylindol-4-yl)-7-(3,4,5-trimethoxyphenyl)-1~{H}-benzimidazole, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | RYX in complex with tubulin
To Be Published
|
|
6LSN
 
 | | Crystal structure of tubulin-inhibitor complex | | Descriptor: | 2-(1-methylindol-5-yl)-7-(3,4,5-trimethoxyphenyl)pyrazolo[1,5-a]pyrimidine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gang, L, Wang, Y.X, Cheng, J.J. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Design, Synthesis, and Bioevaluation of Pyrazolo[1,5-a]Pyrimidine Derivatives as Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site with Potent Anticancer Activities
To Be Published
|
|
1D8V
 
 | | THE RESTRAINED AND MINIMIZED AVERAGE NMR STRUCTURE OF MAP30. | | Descriptor: | ANTI-HIV AND ANTI-TUMOR PROTEIN MAP30 | | Authors: | Wang, Y.-X, Neamati, N, Jacob, J, Palmer, I, Stahl, S.J. | | Deposit date: | 1999-10-26 | | Release date: | 1999-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of anti-HIV-1 and anti-tumor protein MAP30: structural insights into its multiple functions.
Cell(Cambridge,Mass.), 99, 1999
|
|
9IR5
 
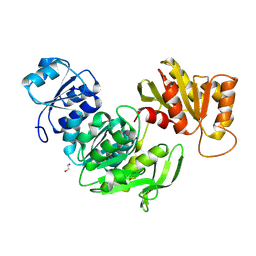 | |
9IR6
 
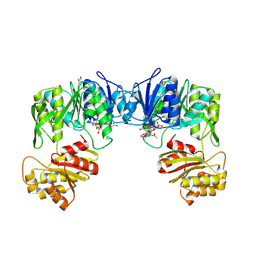 | | Crystal structure of UDP-N-acetylmuramic Acid L-alanine ligase (MurC) from Roseburia faecis in complex with UNAM | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Wang, Y.X, Du, Y.H. | | Deposit date: | 2024-07-14 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Unusual MurC Ligase and Peptidoglycan Discovered in Lachnospiraceae Using a Fluorescent L-Amino Acid Based Selective Labeling Probe.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8WRZ
 
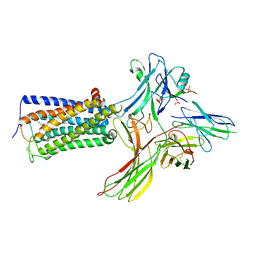 | | Cry-EM structure of cannabinoid receptor-beta-arrestin-1 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
8YIN
 
 | | Cryo-EM structure of Saccharomyces cerevisiae bc1 complex in YF23694-bound state | | Descriptor: | (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, ... | | Authors: | Ye, Y, Li, Z.W, Yang, G.F. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8YHQ
 
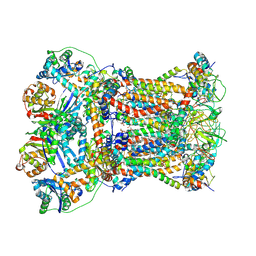 | | Cryo-EM structure of Saccharomyces cerevisiae bc1 complex in pyraclostrobin-bound state | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | Authors: | Ye, Y, Li, Z.W, Yang, G.F. | | Deposit date: | 2024-02-28 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8ZMT
 
 | | Cryo-EM structure of Saccharomyces cerevisiae bc1 complex in Metyltetraprole-bound state | | Descriptor: | (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, ... | | Authors: | Ye, Y, Li, Z.W, Yang, G.F. | | Deposit date: | 2024-05-23 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8YSF
 
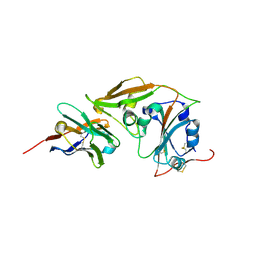 | | MERS-CoV RBD in complex with nanobody Nb9 | | Descriptor: | Nb9, Spike glycoprotein | | Authors: | Wang, Y.X, Ma, S. | | Deposit date: | 2024-03-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure defining of ultrapotent neutralizing nanobodies against MERS-CoV with novel epitopes on receptor binding domain.
Plos Pathog., 20, 2024
|
|
8YSH
 
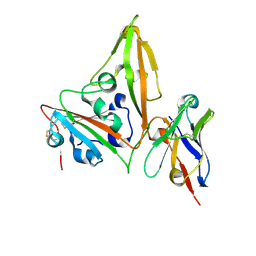 | | MERS-CoV RBD in complex with nanobody Nb14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb14, Spike glycoprotein, ... | | Authors: | Wang, Y.X, Ma, S. | | Deposit date: | 2024-03-23 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure defining of ultrapotent neutralizing nanobodies against MERS-CoV with novel epitopes on receptor binding domain.
Plos Pathog., 20, 2024
|
|
8WYS
 
 | | Local map of human CD5L bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Y.X, Su, C, Xiao, J.Y. | | Deposit date: | 2023-10-31 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | CD5L associates with IgM via the J chain.
Nat Commun, 15, 2024
|
|
8WYR
 
 | | Cryo-EM structure of human CD5L bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Y.X, Su, C, Xiao, J.Y. | | Deposit date: | 2023-10-31 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | CD5L associates with IgM via the J chain.
Nat Commun, 15, 2024
|
|
7CNN
 
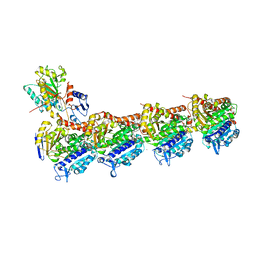 | | vinorelbine in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7CNM
 
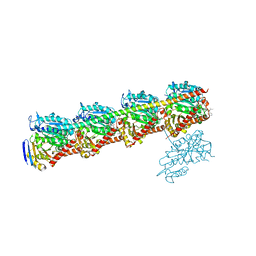 | | YDX in complex with tubulin | | Descriptor: | (2~{S},4~{S})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-4-methyl-3-[[(2~{S},3~{S})-3-methyl-2-[[(2~{R})-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-cyclohexyl-2-methyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
9LHY
 
 | | Crystal structure of a wild-type tagose isomerase (TsT4Ease WT) from Thermotogota bacterium complex with Zn | | Descriptor: | Tagaturonate/fructuronate epimerase, ZINC ION | | Authors: | Wei, H.L, Wang, Y.X, Liu, W.D, Zhu, L.L. | | Deposit date: | 2025-01-13 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Protein Engineering of Tagatose 4-Epimerase for D-Tagatose Production.
J.Agric.Food Chem., 73, 2025
|
|
9LHU
 
 | | Crystal structure of an agose isomerase mutant 5 (TsT4Ease M5) from Thermotogota bacterium with Zn | | Descriptor: | Tagaturonate/fructuronate epimerase, ZINC ION | | Authors: | Wei, H.L, Wang, Y.X, Liu, W.D, Zhu, L.L. | | Deposit date: | 2025-01-13 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Protein Engineering of Tagatose 4-Epimerase for D-Tagatose Production.
J.Agric.Food Chem., 73, 2025
|
|
6WJR
 
 | |
1BVE
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR, 28 STRUCTURES | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1BVG
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1OP1
 
 | | Solution NMR structure of domain 1 of receptor associated protein | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein precursor | | Authors: | Wu, Y, Migliorini, M, Yu, P, Strickland, D.K, Wang, Y.-X. | | Deposit date: | 2003-03-04 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of domain 1 of receptor associated protein.
J.Biomol.Nmr, 26, 2003
|
|
2FTU
 
 | | solution structure of domain 3 of RAP | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein, domain 3 | | Authors: | Lee, D, Walsh, J.D, Wang, Y.-X. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi.
Mol.Cell, 22, 2006
|
|
5YLS
 
 | | Crystal structure of T2R-TTL-Y50 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, E-3-(3-azanyl-4-methoxy-phenyl)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)prop-2-en-1-one, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
5YLJ
 
 | | Crystal structure of T2R-TTL-Millepachine complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxyphenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-17 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
