6J1F
 
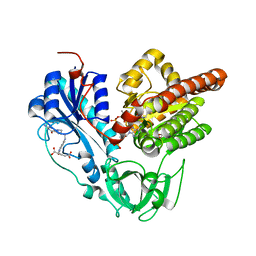 | | Crystal structure of HypX from Aquifex aeolicus in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
6J1I
 
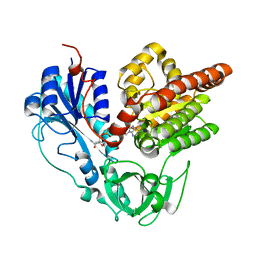 | |
6J1J
 
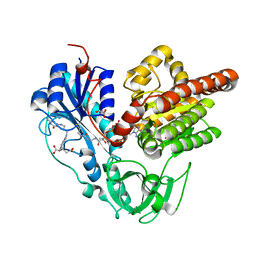 | | Crystal structure of HypX from Aquifex aeolicus, A392F-I419F variant in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
6J1H
 
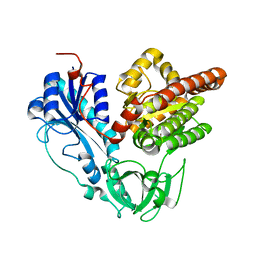 | |
6JIG
 
 | | Crystal structure of GMP reductase C318A from Trypanosoma brucei in complex with guanosine 5'-monophosphate | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Mase, H, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-02-21 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
6JL8
 
 | |
6J0P
 
 | |
6J1E
 
 | |
5H08
 
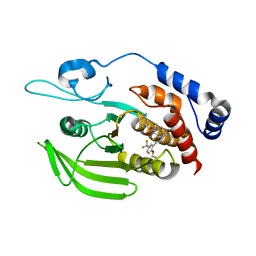 | | Human PTPRZ D1 domain complexed with NAZ2329 | | Descriptor: | 3-{[2-Ethoxy-5-(trifluoromethyl)benzyl]sulfanyl}-N-(phenylsulfonyl)thiophene-2-carboxamide, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2016-10-04 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Targeting PTPRZ inhibits stem cell-like properties and tumorigenicity in glioblastoma cells
Sci Rep, 7, 2017
|
|
2EXV
 
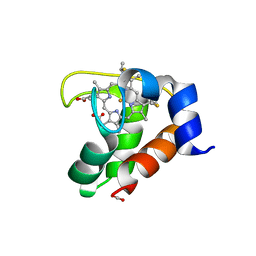 | | Crystal structure of the F7A mutant of the cytochrome c551 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, Cytochrome c-551, HEME C | | Authors: | Bonivento, D, Di Matteo, A, Borgia, A, Travaglini-Allocatelli, C, Brunori, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Unveiling a Hidden Folding Intermediate in c-Type Cytochromes by Protein Engineering
J.Biol.Chem., 281, 2006
|
|
3AL9
 
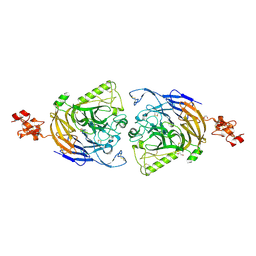 | | Mouse Plexin A2 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2 | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3AFC
 
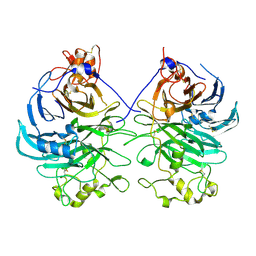 | | Mouse Semaphorin 6A extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin-6A | | Authors: | Yasui, N, Nogi, T, Mihara, E, Takagi, J. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3AL8
 
 | | Plexin A2 / Semaphorin 6A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2, ... | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3T1F
 
 | | Crystal structure of the mouse CD1d-Glc-DAG-s2 complex | | Descriptor: | (2S)-1-(alpha-D-glucopyranosyloxy)-3-(hexadecanoyloxy)propan-2-yl (11Z)-octadec-11-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Girardi, E, Zajonc, D.M. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria.
Nat.Immunol., 12, 2011
|
|
3WPF
 
 | | Crystal structure of mouse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPB
 
 | | Crystal structure of horse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPD
 
 | | Crystal structure of horse TLR9 in complex with inhibitory DNA4084 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*GP*GP*AP*TP*GP*GP*G)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPG
 
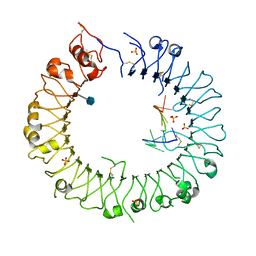 | |
3WPI
 
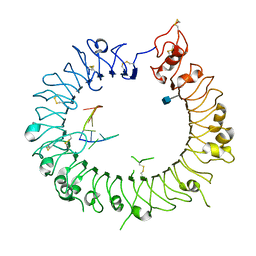 | | Crystal structure of mouse TLR9 in complex with inhibitory DNA_super | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*CP*AP*AP*TP*AP*GP*GP*GP*TP*GP*AP*GP*GP*GP*G)-3'), Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPE
 
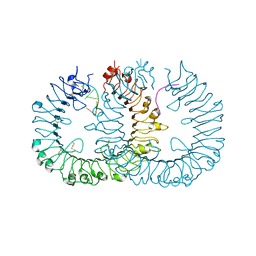 | |
3WCK
 
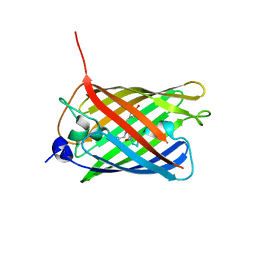 | | Crystal structure of monomeric photosensitizing fluorescent protein, Supernova | | Descriptor: | Monomeric photosenitizing fluorescent protein supernova | | Authors: | Sakai, N, Matsuda, T, Takemoto, K, Nagai, T. | | Deposit date: | 2013-05-27 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation
Sci Rep, 3, 2013
|
|
3WPC
 
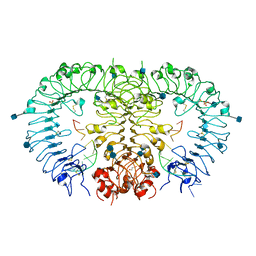 | | Crystal structure of horse TLR9 in complex with agonistic DNA1668_12mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*AP*TP*GP*AP*CP*GP*TP*TP*CP*CP*T)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPH
 
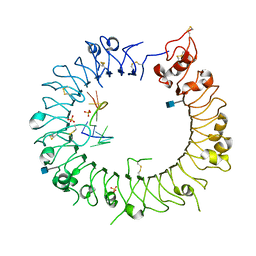 | |
