7FI9
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI7
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI6
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI8
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI5
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
6VKJ
 
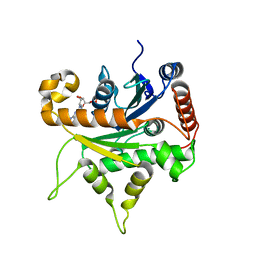 | |
4L5T
 
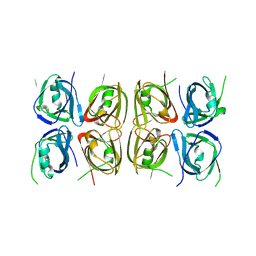 | | Crystal structure of the tetrameric p202 HIN2 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
4L5R
 
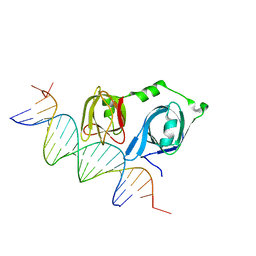 | |
4L5Q
 
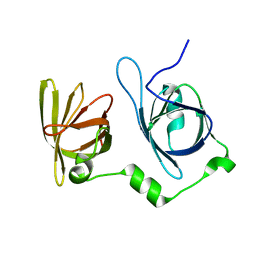 | | Crystal structure of p202 HIN1 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
4L5S
 
 | | p202 HIN1 in complex with 12-mer dsDNA | | Descriptor: | 12-mer DNA, Interferon-activable protein 202, SULFATE ION | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
2GX6
 
 | |
7TXR
 
 | |
7U68
 
 | |
5A3U
 
 | | HIF prolyl hydroxylase 2 (PHD2/EGLN1) in complex with 6-(5-oxo-4-(1H- 1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid | | Descriptor: | 6-(5-oxo-4-(1H-1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid, EGL NINE HOMOLOG 1, MANGANESE (II) ION | | Authors: | Chowdhury, R, Gomez-Perez, V, Schofield, C.J. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Potent and Selective Triazole-Based Inhibitors of the Hypoxia-Inducible Factor Prolyl-Hydroxylases with Activity in the Murine Brain.
Plos One, 10, 2015
|
|
4UWD
 
 | |
5T9W
 
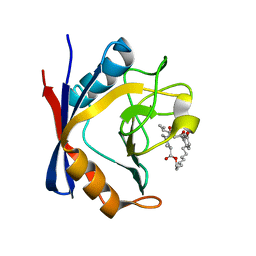 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 5) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 3-[(3-hydroxyphenyl)methyl]-6-(propan-2-yl)-19-oxa-1,4,7,25-tetraazabicyclo[19.3.1]pentacosa-13,15-diene-2,5,8,20-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, P, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T9U
 
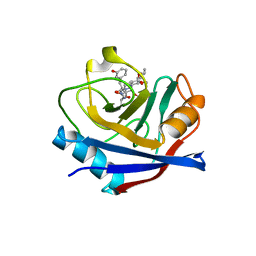 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 3) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 3-[(3-hydroxyphenyl)methyl]-10,12-dimethoxy-9,11-dimethyl-6-(propan-2-yl)-19-oxa-1,4,7,25-tetraazabicyclo[19.3.1]pentacosa-13,15-diene-2,5,8,20-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T9Z
 
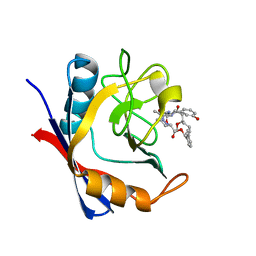 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 6) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 11-[(3-hydroxyphenyl)methyl]-18-methoxy-17-methyl-14-(propan-2-yl)-3-oxa-9,12,15,28-tetraazatricyclo[21.3.1.1~5,9~]octacosa-1(27),21,23,25-tetraene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
1HV9
 
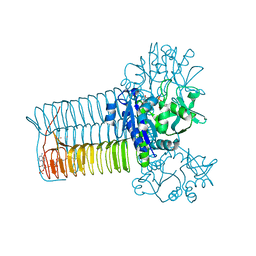 | |
9J6I
 
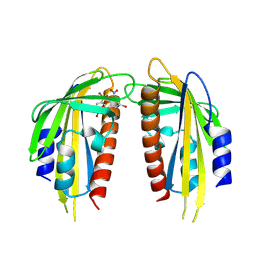 | | Crystal structure of the ABA receptor PYL1 in complex with DBSA compound | | Descriptor: | 5-bromanyl-N2-[(4-bromophenyl)-bis(oxidanyl)-$l^4-sulfanyl]-N1,N1,N3,N3-tetrakis(oxidanyl)benzene-1,2,3-triamine, Abscisic acid receptor PYL1 | | Authors: | Yan, J. | | Deposit date: | 2024-08-16 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stabilization of dimeric PYR/PYL/RCAR family members relieves abscisic acid-induced inhibition of seed germination.
Nat Commun, 15, 2024
|
|
5JZV
 
 | | The structure of D77G hCINAP-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase isoenzyme 6 | | Authors: | Liu, Y, Yang, Z, Yang, Y, Cai, X, Zheng, X. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumour growth.
Nat Commun, 7, 2016
|
|
2LJ2
 
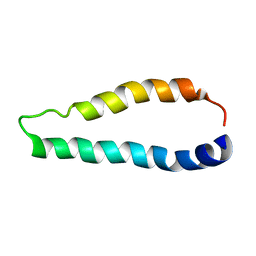 | | Integral membrane core domain of the mercury transporter MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Das, B.B, Nothnagel, H.J, Lu, G.J, Son, W, Park, S, Tian, Y.B, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-09-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of a membrane protein in proteoliposomes.
J.Am.Chem.Soc., 134, 2012
|
|
4DDP
 
 | |
5HMI
 
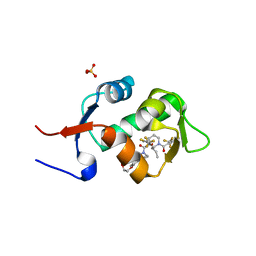 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, {4-[2-(2-hydroxyethoxy)phenyl]piperazin-1-yl}[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]methanone | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HMH
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | 4-[2-(4-{[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]carbonyl}piperazin-1-yl)phenoxy]butanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
