7P6D
 
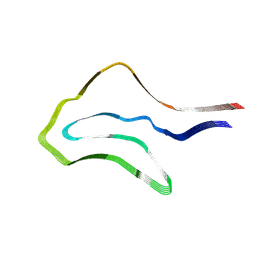 | | Argyrophilic grain disease type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P68
 
 | | Globular glial tauopathy type 3 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P67
 
 | | Globular glial tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6E
 
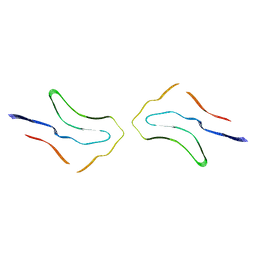 | | Argyrophilic grain disease type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6A
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 1a tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6B
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 1b tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P65
 
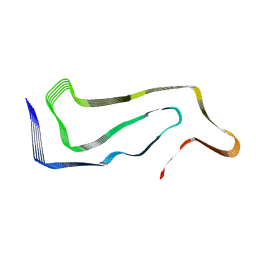 | | Progressive supranuclear palsy tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P66
 
 | | Globular glial tauopathy type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6C
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
5O3O
 
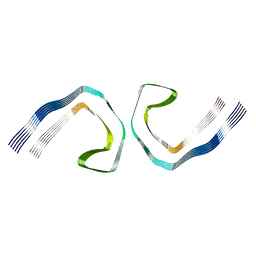 | | Pronase-treated paired helical filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
5NIK
 
 | | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump | | Descriptor: | Macrolide export ATP-binding/permease protein MacB, Macrolide export protein MacA, Outer membrane protein TolC | | Authors: | Fitzpatrick, A.W.P, Llabres, S, Neuberger, A, Blaza, J.N, Bai, X.-C, Okada, U, Murakami, S, van Veen, H.W, Zachariae, U, Scheres, S.H.W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump.
Nat Microbiol, 2, 2017
|
|
5N9Y
 
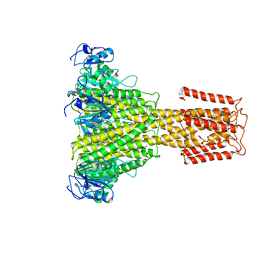 | | The full-length structure of ZntB | | Descriptor: | Zinc transport protein ZntB | | Authors: | Cornelius, G, Stetsenko, A, Scheres, S.H.W, Slotboom, D.J, Guskov, A. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of proton driven zinc transport by ZntB.
Nat Commun, 8, 2017
|
|
5O3L
 
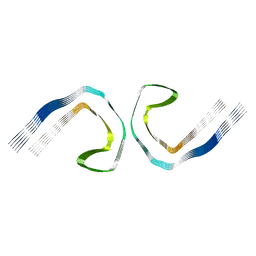 | | Paired helical filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
5O3T
 
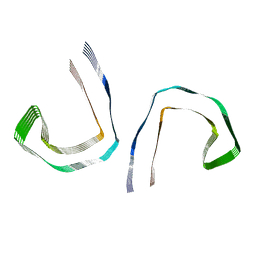 | | Straight filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
5NIL
 
 | | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump-MacB section | | Descriptor: | Macrolide export ATP-binding/permease protein MacB, Macrolide export protein MacA, Outer membrane protein TolC | | Authors: | Fitzpatrick, A.W.P, Llabres, S, Neuberger, A, Blaza, J.N, Bai, X.-C, Okada, U, Murakami, S, van Veen, H.W, Zachariae, U, Scheres, S.H.W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump.
Nat Microbiol, 2, 2017
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
7ARE
 
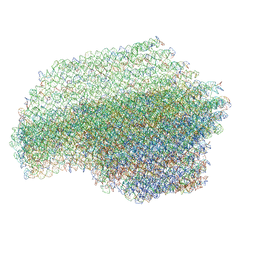 | | DNA origami pointer object v2 | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Thomas, M, Feigl, E, Kohler, F, Kube, M, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7AS5
 
 | | 126 helix bundle DNA nanostructure | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ART
 
 | | 48 helix bundle DNA origami brick | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Feigl, E, Kube, M, Kohler, F, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARV
 
 | | TwistTower_native-twist | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARY
 
 | | Twist-Tower_twist-corrected-variant | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
6ZOY
 
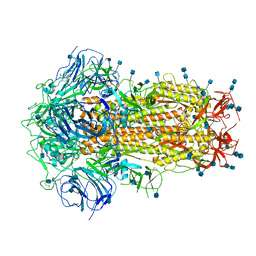 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZWV
 
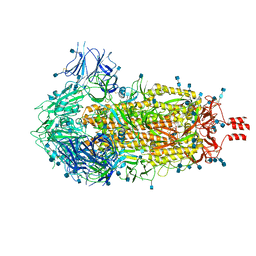 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ke, Z, Qu, K, Nakane, T, Xiong, X, Cortese, M, Zila, V, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Nature, 588, 2020
|
|
6ZOX
 
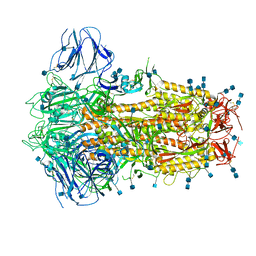 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP2
 
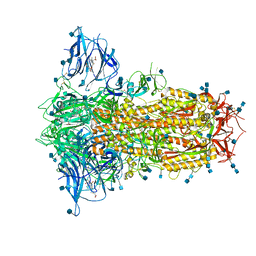 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
