3L25
 
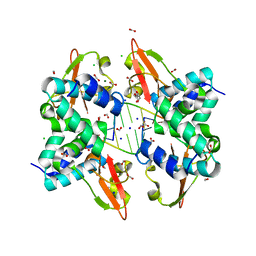 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3P8C
 
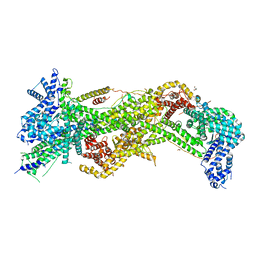 | | Structure and Control of the Actin Regulatory WAVE Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Abl interactor 2, CHLORIDE ION, ... | | Authors: | Chen, Z, Borek, D, Padrick, S.B, Gomez, T.S, Metlagel, Z, Ismail, A.M, Umetani, J, Billadeau, D.D, Otwinowski, Z, Rosen, M.K. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure and control of the actin regulatory WAVE complex.
Nature, 468, 2010
|
|
3L27
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L26
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L28
 
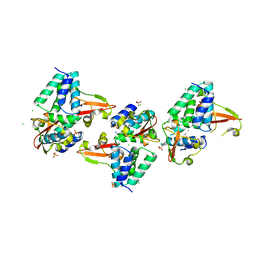 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain K339A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35, SODIUM ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4OFK
 
 | | Crystal Structure of SYG-2 D4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Ozkan, E, Borek, D, Otwinowski, Z, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4IBI
 
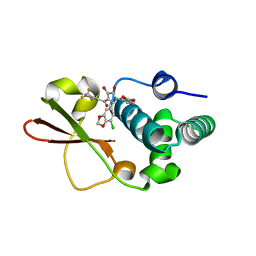 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBJ
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBF
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBE
 
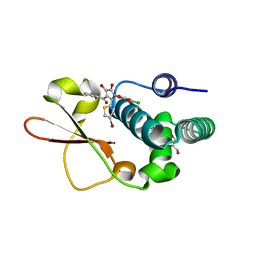 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-chlorobenzoic acid, GLYCEROL, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBK
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBC
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | DIMETHYL SULFOXIDE, Polymerase cofactor VP35, {4-[(2R)-3-(2-chlorobenzoyl)-2-(2-chlorophenyl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBG
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | GLYCEROL, PHOSPHATE ION, Polymerase cofactor VP35, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBD
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBB
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
3KRV
 
 | | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rakonjac, N, Rezacova, P, Borek, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity
To be Published
|
|
3HFR
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Glutamate racemase, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Klimecka, M.M, Cymborowski, M, Skarina, T, Onopriyenko, O, Stam, J, Otwinowski, Z, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes
TO BE PUBLISHED
|
|
3IO4
 
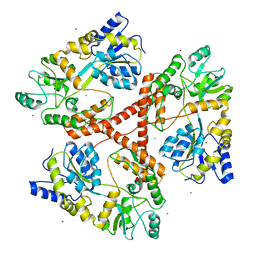 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IO6
 
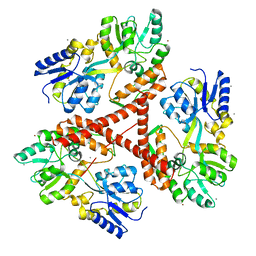 | | Huntingtin amino-terminal region with 17 Gln residues - crystal C92-a | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein, HUNTINGTIN FUSION PROTEIN, ... | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IQ1
 
 | | Crystal structure of DPS protein from Vibrio cholerae O1, a member of a broad superfamily of ferritin-like diiron-carboxylate proteins | | Descriptor: | CHLORIDE ION, DPS family protein | | Authors: | Nocek, B, Peterson, S, Gu, M, Otwinowski, Z, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-18 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of DPS protein from Vibrio cholerae O1, a member of a broad superfamily of ferritin-like diiron-carboxylate proteins
TO BE PUBLISHED
|
|
3IBS
 
 | | Crystal structure of conserved hypothetical protein BatB from Bacteroides thetaiotaomicron | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hattne, J, Bearden, J, Borek, D, Nakka, C, Sather, A, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-16 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of conserved hypothetical protein BatB from Bacteroides thetaiotaomicron
To be Published
|
|
4IO1
 
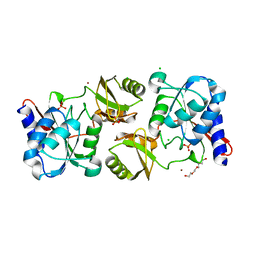 | | Crystal structure of ribose-5-isomerase A from Francisella Tularensis | | Descriptor: | BROMIDE ION, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Rostankowski, R, Nakka, C, Grimshaw, S, Borek, D, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-07 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biophysical studies of Ribose-5-Phosphate Isomerase A from Francisella Tularensis
To be published
|
|
4M8L
 
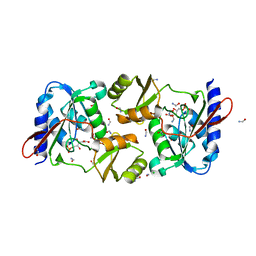 | | crystal structure of RpiA-R5P complex | | Descriptor: | CHLORIDE ION, FORMAMIDE, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Rostankowski, R, Borek, D, Orlikowska, M, Nakka, C, Grimshaw, S, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | crystal structure of RpiA-R5P complex
TO BE PUBLISHED
|
|
3SVA
 
 | | Crystal structure of V57D mutant of human cystatin C | | Descriptor: | ACETATE ION, Cystatin-C, DI(HYDROXYETHYL)ETHER | | Authors: | Orlikowska, M, Szymanska, A, Borek, D, Otwinowski, Z, Skowron, P, Jankowska, E. | | Deposit date: | 2011-07-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural characterization of V57D and V57P mutants of human cystatin C, an amyloidogenic protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHL
 
 | | Structural Basis for Marburg virus VP35 mediate immune evasion mechanisms | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ramanan, P, Borek, D.M, Otwinowski, Z, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for Marburg virus VP35-mediated immune evasion mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
