6N4T
 
 | |
7XGA
 
 | |
3NUY
 
 | |
3NUS
 
 | |
3NUU
 
 | |
3NUN
 
 | |
5TX5
 
 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
3STR
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine hydroxamic acid | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-[2-(hydroxyamino)-2-oxoethyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
3SW8
 
 | |
3SVJ
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine amide | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-methyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-12 | | Release date: | 2011-07-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
7BWB
 
 | | Bombyx mori GH32 beta-fructofuranosidase BmSUC1 | | Descriptor: | Beta-fructofuranosidase | | Authors: | Miyazaki, T, Oba, N. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the substrate specificity of Bombyx mori beta-fructofuranosidase belonging to the glycoside hydrolase family 32.
Insect Biochem.Mol.Biol., 127, 2020
|
|
7BWC
 
 | |
1J0F
 
 | | Solution Structure of the SH3 Domain Binding Glutamic Acid-rich Protein Like 3 | | Descriptor: | SH3 domain-binding glutamic acid-rich-like protein 3 | | Authors: | Miyamoto, K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the SH3 Domain Binding Glutamic Acid-rich Protein Like 3
To be Published
|
|
1J0G
 
 | | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold | | Descriptor: | Hypothetical Protein 1810045K17 | | Authors: | Zhao, C, Kigawa, T, Koshiba, S, Tochio, N, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold
To be Published
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
7VH9
 
 | | Solution structure of the chimeric peptide of the first SURP domain of Human SF3A1 and the interacting region of SF1. | | Descriptor: | Splicing factor 3A subunit 1,Splicing factor 1 | | Authors: | Muto, Y, Kuwasako, K, Takizawa, M, Kobayashi, N, Sakamoto, T. | | Deposit date: | 2021-09-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction between the first SURP domain of the SF3A1 subunit in U2 snRNP and the human splicing factor SF1.
Protein Sci., 31, 2022
|
|
4RS3
 
 | | Crystal structure of carbohydrate transporter A0QYB3 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with xylitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4RSM
 
 | | Crystal structure of carbohydrate transporter msmeg_3599 from mycobacterium smegmatis str. mc2 155, target efi-510970, in complex with d-threitol | | Descriptor: | D-Threitol, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
7BV9
 
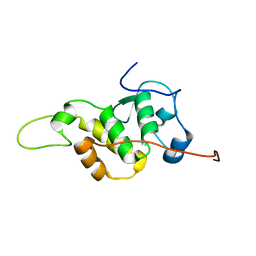 | | The NMR structure of the BEN domain from human NAC1 | | Descriptor: | Nucleus accumbens-associated protein 1 | | Authors: | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
6R5F
 
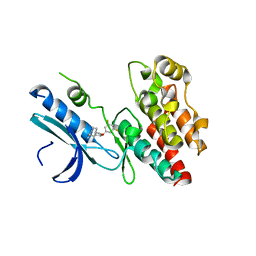 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
7DNS
 
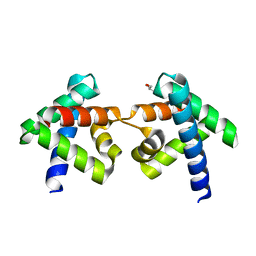 | | Crystal structure of domain-swapped dimer of H5_Fold-0 Elsa; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Suzuki, K, Kobayashi, N, Murata, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
4N4U
 
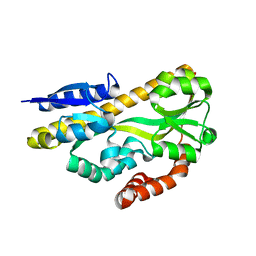 | | Crystal structure of ABC transporter solute binding protein BB0719 from Bordetella bronchiseptica RB50, TARGET EFI-510049 | | Descriptor: | GLYCEROL, Putative ABC transporter periplasmic solute-binding protein | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
