6CWX
 
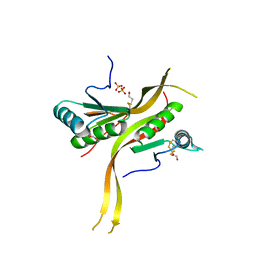 | | Crystal structure of human ribonuclease P/MRP proteins Rpp20/Rpp25 | | Descriptor: | FORMIC ACID, Ribonuclease P protein subunit p20, Ribonuclease P protein subunit p25, ... | | Authors: | Chan, C.W, Kiesel, B.R, Mondragon, A. | | Deposit date: | 2018-03-31 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rpp20/Rpp25 Reveals Quaternary Level Adaptation of the Alba Scaffold as Structural Basis for Single-stranded RNA Binding.
J. Mol. Biol., 430, 2018
|
|
6UGG
 
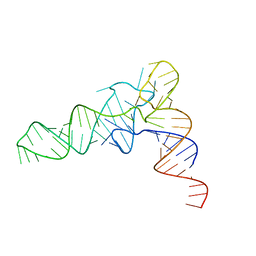 | | Structure of unmodified E. coli tRNA(Asp) | | Descriptor: | tRNAasp | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs.
Rna, 26, 2020
|
|
6UGI
 
 | |
6VMY
 
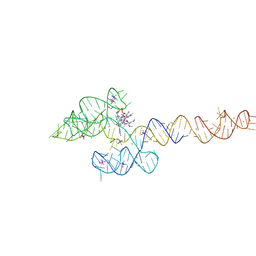 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
1OIS
 
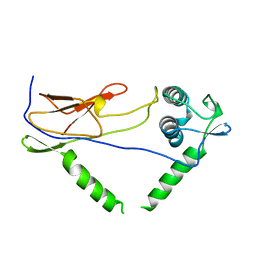 | | YEAST DNA TOPOISOMERASE I, N-TERMINAL FRAGMENT | | Descriptor: | DNA TOPOISOMERASE I | | Authors: | Lue, N, Sharma, A, Mondragon, A, Wang, J.C. | | Deposit date: | 1996-09-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 26 kDa yeast DNA topoisomerase I fragment: crystallographic structure and mechanistic implications.
Structure, 3, 1995
|
|
1CYY
 
 | |
1CY9
 
 | |
6N1R
 
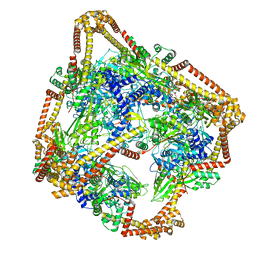 | | Tetrahedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in tetrahedral symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6NBT
 
 | |
6N1Q
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in D2 symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6N1P
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment with DNA, solved by cryoEM in C2 symmetry | | Descriptor: | DNA (44-MER), DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.35 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6NBU
 
 | |
2A2E
 
 | | Crystal structure of the RNA subunit of Ribonuclease P. Bacterial A-type. | | Descriptor: | OSMIUM ION, RNA subunit of RNase P | | Authors: | Torres-Larios, A, Swinger, K.K, Krasilnikov, A.S, Pan, T, Mondragon, A. | | Deposit date: | 2005-06-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the RNA component of bacterial ribonuclease P.
Nature, 437, 2005
|
|
1CUN
 
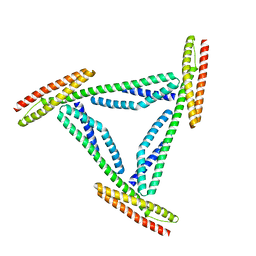 | | CRYSTAL STRUCTURE OF REPEATS 16 AND 17 OF CHICKEN BRAIN ALPHA SPECTRIN | | Descriptor: | PROTEIN (ALPHA SPECTRIN) | | Authors: | Grum, V.L, Li, D, MacDonald, R.I, Mondragon, A. | | Deposit date: | 1999-08-20 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two repeats of spectrin suggest models of flexibility.
Cell(Cambridge,Mass.), 98, 1999
|
|
1NBS
 
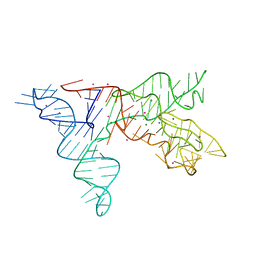 | | Crystal structure of the specificity domain of Ribonuclease P RNA | | Descriptor: | LEAD (II) ION, MAGNESIUM ION, RIBONUCLEASE P RNA | | Authors: | Krasilnikov, A.S, Yang, X, Pan, T, Mondragon, A. | | Deposit date: | 2002-12-03 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the specificity domain of Ribonuclease P
Nature, 421, 2003
|
|
1M63
 
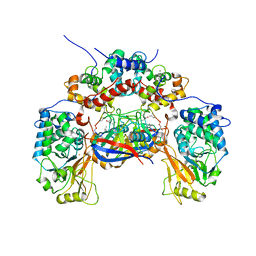 | | Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes | | Descriptor: | CALCINEURIN B SUBUNIT ISOFORM 1, CALCIUM ION, CYCLOSPORIN A, ... | | Authors: | Huai, Q, Kim, H.-Y, Liu, Y, Zhao, Y, Mondragon, A, Liu, J.O, Ke, H. | | Deposit date: | 2002-07-12 | | Release date: | 2002-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Calcineurin-Cyclophilin-Cyclosporin Shows Common But Distinct Recognition of Immunophilin-Drug Complexes
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2O59
 
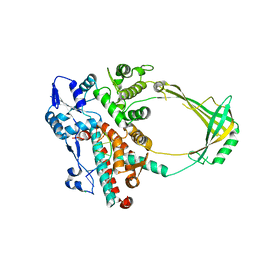 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol pH 8.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O5C
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O19
 
 | | Structure of E. coli topoisomersae III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R, Mondragon, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O54
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, Digate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O5E
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
3M4A
 
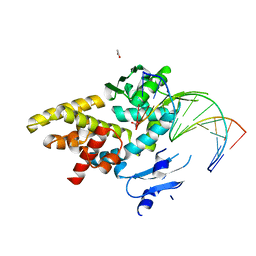 | | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site | | Descriptor: | ACETIC ACID, DNA (5'-D(*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*CP*CP*TP*TP*AP*TP*TP*C)-3'), ... | | Authors: | Patel, A, Yakovleva, L, Shuman, S, Mondragon, A. | | Deposit date: | 2010-03-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site.
Structure, 18, 2010
|
|
3F57
 
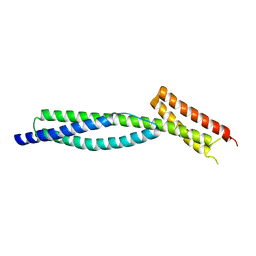 | |
3F59
 
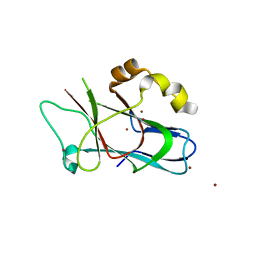 | |
1Q08
 
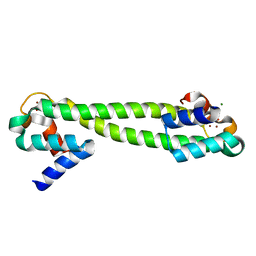 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator, at 1.9 A resolution (space group P212121) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
