7L3R
 
 | |
5VXB
 
 | |
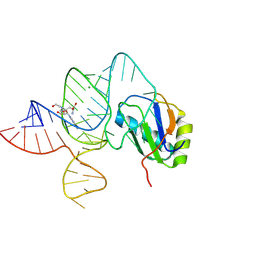
6LAX
 
 | | the mutant SAM-VI riboswitch (U6C) bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Sun, A, Ren, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
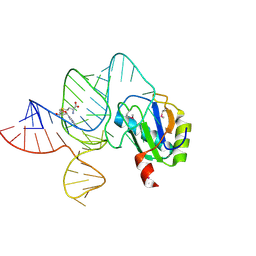
6LAZ
 
 | | the wildtype SAM-VI riboswitch bound to a N-mustard SAM analog M1 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-hydroxyethyl)amino]-2-azaniumyl-butanoate, MAGNESIUM ION, RNA (55-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LAS
 
 | | the wildtype SAM-VI riboswitch bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
3NCU
 
 | |
8GXB
 
 | |
8GXC
 
 | |
6ZXZ
 
 | | Sarcin-Ricin Loop RNA from Ecoli with a A2670-2'-OCF3 modification | | Descriptor: | POTASSIUM ION, SODIUM ION, Sarcin-Ricin Loop RNA from Ecoli with a A2670-2'-OCF3 modification | | Authors: | Ennifar, E. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | 2'- O -Trifluoromethylated RNA - a powerful modification for RNA chemistry and NMR spectroscopy.
Chem Sci, 11, 2020
|
|
6ZYB
 
 | |
9F3F
 
 | | Trypanosoma brucei nuclear cap-binding complex (CBC) bound to cap0 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, Nuclear cap binding complex subunit CBP110, Nuclear cap binding complex subunit CBP30, ... | | Authors: | Bernhard, H, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2024-04-25 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of Spliced Leader RNA recognition by the Trypanosoma brucei cap-binding complex.
Nat Commun, 16, 2025
|
|
9F67
 
 | | Trypanosoma brucei nuclear cap-binding complex (CBC) bound to cap4 | | Descriptor: | Nuclear cap binding complex subunit CBP110, Nuclear cap binding complex subunit CBP30, Nuclear cap-binding protein subunit 2, ... | | Authors: | Bernhard, H, Warminski, M, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2024-04-30 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of Spliced Leader RNA recognition by the Trypanosoma brucei cap-binding complex.
Nat Commun, 16, 2025
|
|
4RGF
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
5TGA
 
 | |
5TGM
 
 | | Crystal structure of the S.cerevisiae 80S ribosome in complex with the A-site bound aminoacyl-tRNA analog ACCA-Pro | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Melnikov, S, Mailliot, J, Yusupov, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular insights into protein synthesis with proline residues.
EMBO Rep., 17, 2016
|
|
5D99
 
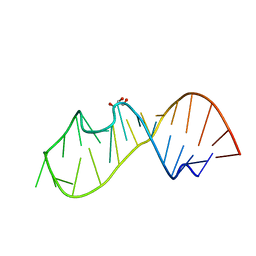 | | 3DW4 redetermined by direct methods starting from random phase angles | | Descriptor: | GLYCEROL, RNA (27-MER) hairpin from sarcin-ricin domain of E. coli 23S rRNA | | Authors: | Mooers, B.H.M. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6JQ5
 
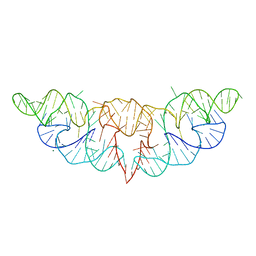 | | The structure of Hatchet Ribozyme | | Descriptor: | MAGNESIUM ION, RNA (82-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JQ6
 
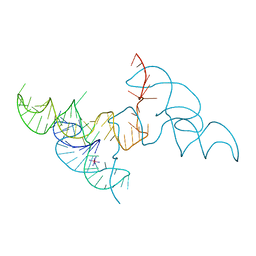 | | Hatchet Ribozyme Structure soaking with Ir(NH3)6+ | | Descriptor: | IRIDIUM HEXAMMINE ION, RNA (81-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1Y27
 
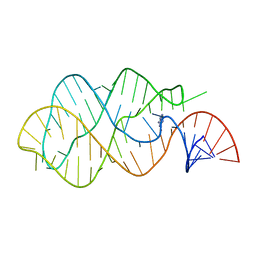 | | G-riboswitch-guanine complex | | Descriptor: | Bacillus subtilis xpt, GUANINE | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
6LAU
 
 | | the wildtype SAM-VI riboswitch bound to SAH | | Descriptor: | CESIUM ION, GUANOSINE-5'-TRIPHOSPHATE, RNA (54-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
1Y26
 
 | | A-riboswitch-adenine complex | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus A-riboswitch | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
5K7C
 
 | | The native structure of native pistol ribozyme | | Descriptor: | DNA/RNA 11-MER, MAGNESIUM ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5K7D
 
 | | The structure of native pistol ribozyme, bound to Iridium | | Descriptor: | DNA/RNA 11-MER, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5LYB
 
 | |
