2G8N
 
 | | Structure of hPNMT with inhibitor 3-Hydroxymethyl-7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | (3R)-N-(4-CHLOROPHENYL)-3-(HYDROXYMETHYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Gee, C.L, Martin, J.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of the Binding of 3-Fluoromethyl-7-sulfonyl-1,2,3,4-tetrahydroisoquinolines with Their Isosteric Sulfonamides to the Active Site of Phenylethanolamine N-Methyltransferase
J.Med.Chem., 49, 2006
|
|
2G70
 
 | | Structure of human PNMT in complex with inhibitor 3-hydroxymethyl-7-nitro-THIQ and AdoMet (SAM) | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Tyndall, J.D.A, Gee, C.L, Martin, J.L. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
2G71
 
 | | Structure of hPNMT with inhibitor 3-fluoromethyl-7-trifluoropropyl-THIQ and AdoHcy | | Descriptor: | (3R)-3-(FLUOROMETHYL)-N-(3,3,3-TRIFLUOROPROPYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, GLYCEROL, Phenylethanolamine N-methyltransferase, ... | | Authors: | Tyndall, J.D.A, Gee, C.L, Martin, J.L. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
1KNG
 
 | | Crystal structure of CcmG reducing oxidoreductase at 1.14 A | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN CYCY | | Authors: | Edeling, M.A, Guddat, L.W, Fabianek, R.A, Thony-Meyer, L, Martin, J.L. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment
Structure, 10, 2002
|
|
2D06
 
 | | Human Sult1A1 Complexed With Pap and estradiol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ESTRADIOL, Sulfotransferase 1A1 | | Authors: | Gamage, N.U, Tsvetanov, S, Duggleby, R.G, McManus, M.E, Martin, J.L. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of human SULT1A1 crystallized with estradiol. An insight into active site plasticity and substrate inhibition with multi-ring substrates
J.Biol.Chem., 280, 2005
|
|
1KQU
 
 | | Human phospholipase A2 complexed with a substrate anologue | | Descriptor: | 6-PHENYL-4(R)-(7-PHENYL-HEPTANOYLAMINO)-HEXANOIC ACID, CALCIUM ION, Phospholipase A2, ... | | Authors: | Tyndall, J.D, Martin, J.L. | | Deposit date: | 2002-01-07 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D-Tyrosine as a chiral precusor to potent inhibitors of human nonpancreatic secretory phospholipase A2 (IIa) with antiinflammatory activity.
Chembiochem, 4, 2003
|
|
2AN3
 
 | | Structure of PNMT with S-adenosyl-L-homocysteine and the semi-rigid analogue acceptor substrate cis-(1R,2S)-2-amino-1-tetralol. | | Descriptor: | CIS-(1R,2S)-2-AMINO-1,2,3,4-TETRAHYDRONAPHTHALEN-1-OL, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2AN5
 
 | | Structure of human PNMT complexed with S-adenosyl-homocysteine and an inhibitor, trans-(1S,2S)-2-amino-1-tetralol | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2AN4
 
 | | Structure of PNMT complexed with S-adenosyl-L-homocysteine and the acceptor substrate octopamine | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
1ACV
 
 | | DSBA MUTANT H32S | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
1AC1
 
 | | DSBA MUTANT H32L | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
4ZL7
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal I | | Descriptor: | HEXAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL8
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahoh, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL9
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal III | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
1D4L
 
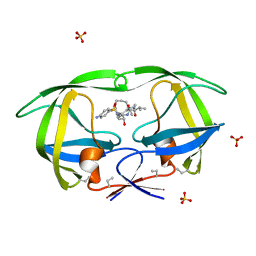 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1D4K
 
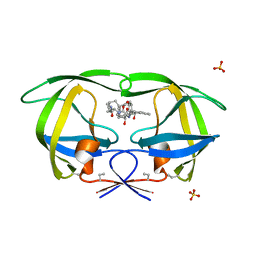 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1NUL
 
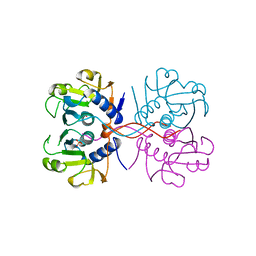 | | XPRTASE FROM E. COLI | | Descriptor: | MAGNESIUM ION, SULFATE ION, XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1996-10-15 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli xanthine phosphoribosyltransferase.
Biochemistry, 36, 1997
|
|
6BR4
 
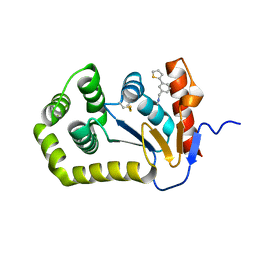 | | Crystal structure of Escherichia coli DsbA in complex with {N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
3PUK
 
 | | Re-refinement of the crystal structure of Munc18-3 and Syntaxin4 N-peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 3 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1V58
 
 | | Crystal Structure Of the Reduced Protein Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V57
 
 | | Crystal Structure of the Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3KPW
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 1-Aminoisoquinoline | | Descriptor: | ISOQUINOLIN-1-AMINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQP
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Aminoquinoline | | Descriptor: | Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, quinolin-6-amine | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQT
 
 | |
3KPU
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-quinolinol | | Descriptor: | Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, quinolin-4-ol | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
