7VSP
 
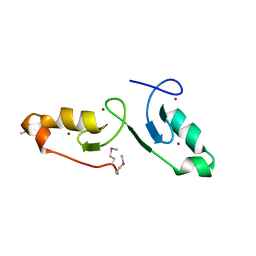 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
7VSI
 
 | | Structure of human SGLT2-MAP17 complex bound with empagliflozin | | Descriptor: | (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[[4-[(3S)-oxolan-3-yl]oxyphenyl]methyl]phenyl]-6-(hydroxymethyl)oxane-3,4,5-triol, PALMITIC ACID, PDZK1-interacting protein 1, ... | | Authors: | Chen, L, Niu, Y, Liu, R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of inhibition of the human SGLT2-MAP17 glucose transporter.
Nature, 601, 2022
|
|
7XQV
 
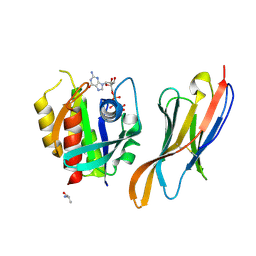 | | The complex of nanobody Rh57 binding to GTP-bound RhoA active form | | Descriptor: | ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhang, Y.R, Liu, R, Ding, Y. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insights into the binding of nanobody Rh57 to active RhoA-GTP.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
1PFN
 
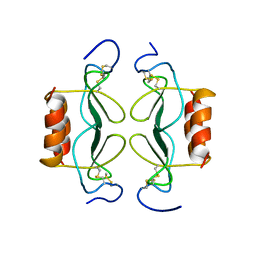 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1PFM
 
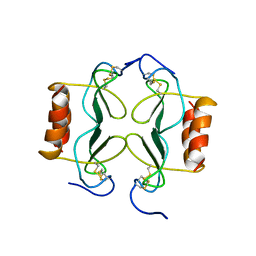 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
5JVA
 
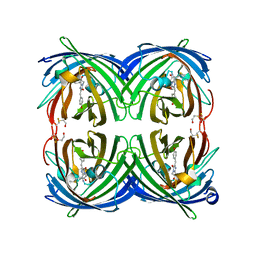 | | 1.95 angstrom crystal structure of TAGRFP-T | | Descriptor: | BETA-MERCAPTOETHANOL, SULFATE ION, TagRFP-T | | Authors: | Hu, X.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of red fluorescent protein TagRFP-T reveals the mechanism of its superior photostability.
Biochem. Biophys. Res. Commun., 477, 2016
|
|
5OEP
 
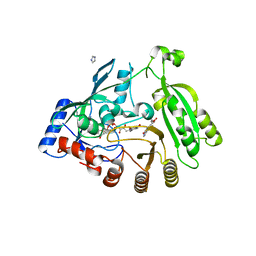 | | Mycobacterium tuberculosis DprE1 in complex with inhibitor TCA481 | | Descriptor: | Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2017-07-09 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OEL
 
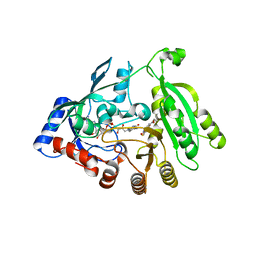 | | Mycobacterium tuberculosis DprE1 mutant Y314C in complex with TCA1 | | Descriptor: | Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ethyl ({2-[(1,3-benzothiazol-2-ylcarbonyl)amino]thiophen-3-yl}carbonyl)carbamate | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2017-07-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OEQ
 
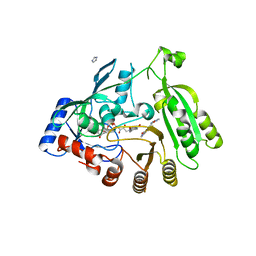 | | Mycobacterium tuberculosis DprE1 in complex with inhibitor TCA020 | | Descriptor: | Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2017-07-09 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8Z02
 
 | | CoA bound to human GTP-specific succinyl-CoA synthetase | | Descriptor: | COENZYME A, PHOSPHATE ION, Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, ... | | Authors: | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
8Z03
 
 | | Lactate bound to human GTP-specific succinyl-CoA synthetase | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, COENZYME A, MAGNESIUM ION, ... | | Authors: | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
8YGG
 
 | | pP1192R-apo Closed state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YGH
 
 | | pP1192R-apo open state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YGE
 
 | | pP1192R-DNA-m-AMSA complex DNA binding/cleavage domain | | Descriptor: | DNA (12-mer), DNA (20-mer), DNA (8-mer), ... | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YIK
 
 | | pP1192R-ATPase-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
3I19
 
 | |
6LMH
 
 | |
5W0C
 
 | | Cytochrome P450 (CYP) 2C9 TCA007 Inhibitor Complex | | Descriptor: | Cytochrome P450 2C9, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnson, E.F, Hsu, M.-H. | | Deposit date: | 2017-05-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6Z17
 
 | | PqsR (MvfR) in complex with antagonist 6 | | Descriptor: | 6-chloranyl-3-[(2-propan-2-yl-2,3-dihydro-1,3-thiazol-4-yl)methyl]quinazolin-4-one, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
6YZ3
 
 | | PqsR (MvfR) in complex with antagonist 19 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-[(2-hexyl-2,3-dihydro-1,3-thiazol-4-yl)methyl]quinazolin-4-one, LysR family transcriptional regulator | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
6Z07
 
 | | PqsR (MvfR) in complex with antagonist 12 | | Descriptor: | 3-[(2-~{tert}-butyl-1,3-thiazol-4-yl)methyl]-6-chloranyl-quinazolin-4-one, GLYCEROL, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
6TPR
 
 | | PqsR (MvfR) bound to inhibitory compound 40 | | Descriptor: | 2-[(5-methyl-[1,2,4]triazino[5,6-b]indol-3-yl)sulfanyl]-~{N}-(4-pyridin-2-yloxyphenyl)ethanamide, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm GrownPseudomonas aeruginosa.
Front Chem, 8, 2020
|
|
9JDU
 
 | | The crystal structure of PDE10A complexed with inhibitor 2061 | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2024-09-01 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.3000176 Å) | | Cite: | Discovery of novel azetidine-based imidazopyridines as selective and orally bioavailable inhibitors of phosphodiesterase 10A for the treatment of pulmonary arterial hypertension.
Eur.J.Med.Chem., 290, 2025
|
|
8R19
 
 | | SARS-CoV-2 Mpro (Omicron, P132H) free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R0V
 
 | | SARS-CoV-2 Mpro (Omicron, P132H) in complex with alpha-ketoamide 13b-K at pH 6.5 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
