3FW4
 
 | |
8CWY
 
 | |
8CUS
 
 | |
8CWS
 
 | |
8CUT
 
 | |
8CUX
 
 | |
8CWZ
 
 | |
8CUU
 
 | |
8CUV
 
 | |
8CUW
 
 | |
6THG
 
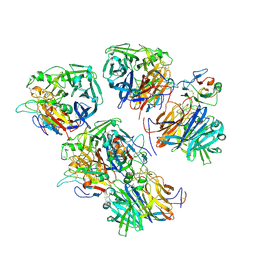 | | Cedar Virus attachment glycoprotein (G) in complex with human ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Pryce, R, Rissanen, I, Harlos, K, Bowden, T. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.074 Å) | | Cite: | A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus.
Life Sci Alliance, 3, 2020
|
|
6THB
 
 | | Receptor binding domain of the Cedar Virus attachment glycoprotein (G) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | Authors: | Pryce, R, Rissanen, I, Harlos, K, Bowden, T. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus.
Life Sci Alliance, 3, 2020
|
|
8DQL
 
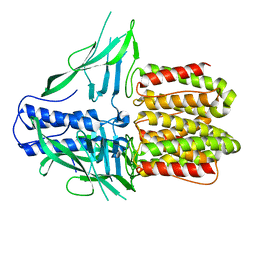 | | CryoEM structure of IglD | | Descriptor: | Secretion system protein | | Authors: | Liu, X, Clemens, D, Lee, B, Yang, X, Zhou, H, Horwitz, M. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic Structure of IglD Demonstrates Its Role as a Component of the Baseplate Complex of the Francisella Type VI Secretion System.
Mbio, 13, 2022
|
|
8CKF
 
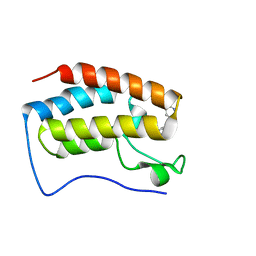 | | Crystal Structure of the first bromodomain of human BRD4 L94C variant in complex with racemic 3,5-dimethylisoxazol ligand | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{S})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Thomas, A.M, McDonough, M.A, Schiedel, M, Conway, S.J. | | Deposit date: | 2023-02-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutate and Conjugate: A Method to Enable Rapid In-Cell Target Validation.
Acs Chem.Biol., 18, 2023
|
|
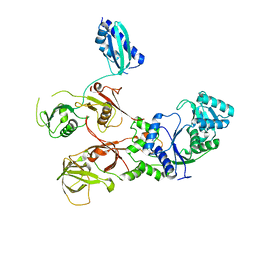
7WRS
 
 | |
7WRU
 
 | |
4ZJV
 
 | |
6JKG
 
 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
6JKH
 
 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
7W3D
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Descriptor: | Bromodomain-containing protein 4, N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of BET specific bromodomain inhibitors with a novel scaffold.
Bioorg.Med.Chem., 72, 2022
|
|
4OK0
 
 | | Crystal structure of putative nucleotidyltransferase from H. pylori | | Descriptor: | Putative | | Authors: | Yoon, J.Y, Lee, S.J, Lee, B, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-01-21 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of JHP933 from Helicobacter pylori J99 shows two-domain architecture with a DUF1814 family nucleotidyltransferase domain and a helical bundle domain.
Proteins, 82, 2014
|
|
2KI2
 
 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | Descriptor: | Ss-DNA binding protein 12RNP2 | | Authors: | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
8SZZ
 
 | |
8FAR
 
 | |
5H5M
 
 | | Crystal structure of HMP-1 M domain | | Descriptor: | Alpha-catenin-like protein hmp-1 | | Authors: | Kang, H, Bang, I, Weis, W.I, Choi, H.J. | | Deposit date: | 2016-11-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of Caenorhabditis elegans alpha-catenin reveals constitutive binding to beta-catenin and F-actin
J. Biol. Chem., 292, 2017
|
|
