5OWJ
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 2) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
1ZRY
 
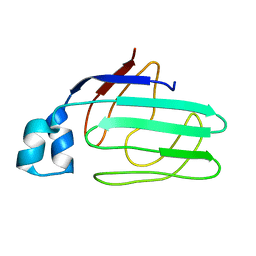 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
1ZT3
 
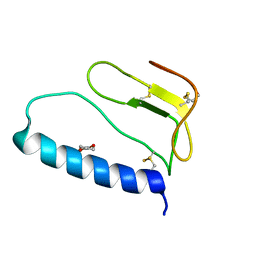 | | C-terminal domain of Insulin-like Growth Factor Binding Protein-1 isolated from human amniotic fluid | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Insulin-like growth factor binding protein 1 | | Authors: | Sala, A, Capaldi, S, Campagnoli, M, Faggion, B, Labo, S, Perduca, M, Romano, A, Carrizo, M.E, Valli, M, Visai, L, Minchiotti, L, Galliano, M, Monaco, H.L. | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Properties of the C-terminal Domain of Insulin-like Growth Factor-binding Protein-1 Isolated from Human Amniotic Fluid
J.Biol.Chem., 280, 2005
|
|
5OTO
 
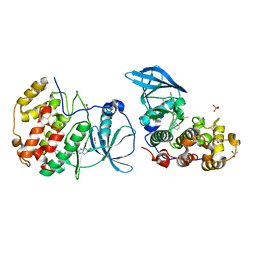 | | The crystal structure of CK2alpha in complex with compound 30 | | Descriptor: | 2-(5-chloranyl-1~{H}-benzimidazol-2-yl)-~{N}-[[3-chloranyl-4-(2-ethylphenyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1ZT5
 
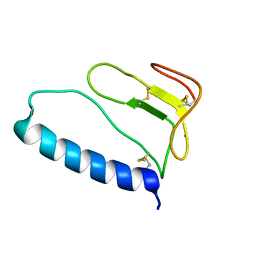 | | C-terminal domain of Insulin-like Growth Factor Binding Protein-1 isolated from human amniotic fluid complexed with Iron(II) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (II) ION, Insulin-like growth factor binding protein 1 | | Authors: | Sala, A, Capaldi, S, Campagnoli, M, Faggion, B, Labo, S, Perduca, M, Romano, A, Carrizo, M.E, Valli, M, Visai, L, Minchiotti, L, Galliano, M, Monaco, H.L. | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structure and Properties of the C-terminal Domain of Insulin-like Growth Factor-binding Protein-1 Isolated from Human Amniotic Fluid
J.Biol.Chem., 280, 2005
|
|
1H15
 
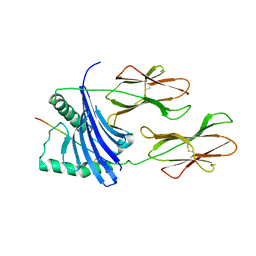 | | X-ray crystal structure of HLA-DRA1*0101/DRB5*0101 complexed with a peptide from Epstein Barr Virus DNA polymerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA POLYMERASE, ... | | Authors: | Lang, H, Jacobsen, H, Ikemizu, S, Andersson, C, Harlos, K, Madsen, L, Hjorth, P, Sondergaard, L, Svejgaard, A, Wucherpfennig, K, Stuart, D.I, Bell, J.I, Jones, E.Y, Fugger, L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Functional and Structural Basis for Tcr Cross-Reactivity in Multiple Sclerosis
Nat.Immunol., 3, 2002
|
|
8CEA
 
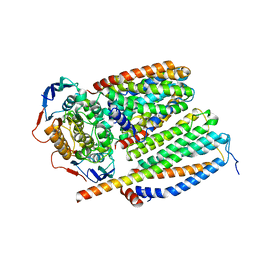 | | Cytochrome c maturation complex CcmABCD, E154Q | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CE1
 
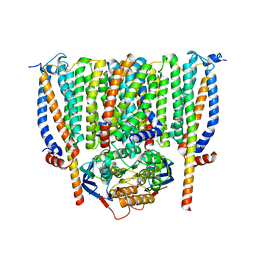 | | Cytochrome c maturation complex CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CE5
 
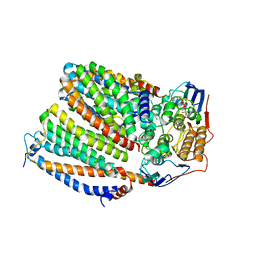 | | Cytochrome c maturation complex CcmABCD, E154Q, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CE8
 
 | | Cytochrome c maturation complex CcmABCDE | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Cytochrome c-type biogenesis protein CcmE, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
3JXU
 
 | | Crystal structure of the human 70kDa heat shock protein 1A (Hsp70-1) ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1, MAGNESIUM ION, ... | | Authors: | Wisniewska, M.M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
3K1R
 
 | |
3JAX
 
 | | Heavy meromyosin from Schistosoma mansoni muscle thick filament by negative stain EM | | Descriptor: | myosin 2 heavy chain, myosin regulatory light chain, smooth muscle myosin essential light chain | | Authors: | Sulbaran, G, Alamo, L, Pinto, A, Marquez, G, Mendez, F, Padron, R, Craig, R. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | An invertebrate smooth muscle with striated muscle myosin filaments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8C1V
 
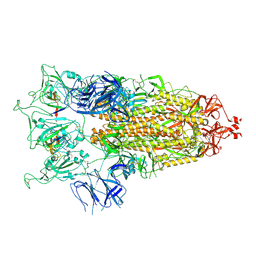 | | SARS-CoV-2 S-trimer (3 RBDs up) bound to TriSb92, fitted into cryo-EM map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sb92, ... | | Authors: | Huiskonen, J.T, Rissanen, I, Hannula, L. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Intranasal trimeric sherpabody inhibits SARS-CoV-2 including recent immunoevasive Omicron subvariants.
Nat Commun, 14, 2023
|
|
3K1W
 
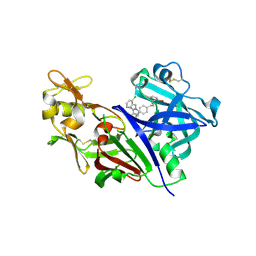 | | New Classes of Potent and Bioavailable Human Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetylamino-2-deoxy-alpha-L-idopyranose, 4-{4-[3-(2-bromo-5-fluorophenoxy)propyl]phenyl}-N-(2-chlorobenzyl)-N-cyclopropyl-1,2,5,6-tetrahydropyridine-3-carboxamide, ... | | Authors: | Prade, L. | | Deposit date: | 2009-09-29 | | Release date: | 2010-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New classes of potent and bioavailable human renin inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5JCJ
 
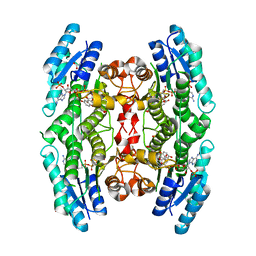 | | Trypanosoma brucei PTR1 in complex with inhibitor NMT-H037 (compound 7) | | Descriptor: | 2-(3,4-dihydroxyphenyl)-3,6-dihydroxy-4H-1-benzopyran-4-one, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Landi, G, Pozzi, C, Di Pisa, F, Dello Iacono, L, Mangani, S. | | Deposit date: | 2016-04-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Profiling of Flavonol Derivatives for the Development of Antitrypanosomatidic Drugs.
J.Med.Chem., 59, 2016
|
|
4MER
 
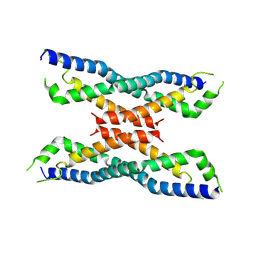 | | Crystal structure of the novel protein and virulence factor sHIP (Q99XU0) from Streptococcus pyogenes | | Descriptor: | streptococcal Histidine-rich glycoprotein Interacting Protein | | Authors: | Wisniewska, M, Happonen, L, Frick, M.-I, Bjorck, L, Streicher, W, Malmstrom, J, Wikstrom, M. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional and Structural Properties of a Novel Protein and Virulence Factor (Protein sHIP) in Streptococcus pyogenes.
J.Biol.Chem., 289, 2014
|
|
5JDC
 
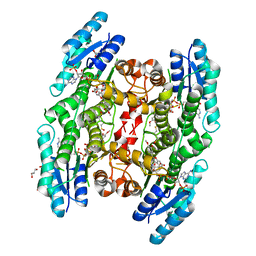 | | Trypanosoma brucei PTR1 in complex with inhibitor NP-13 (Hesperetin) | | Descriptor: | (2S)-5,7-dihydroxy-2-(3-hydroxy-4-methoxyphenyl)-2,3-dihydro-4H-1-benzopyran-4-one, ACETATE ION, GLYCEROL, ... | | Authors: | Mangani, S, Pozzi, C, Di Pisa, F, Landi, G, Dello Iacono, L. | | Deposit date: | 2016-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Profiling of Flavonol Derivatives for the Development of Antitrypanosomatidic Drugs.
J.Med.Chem., 59, 2016
|
|
3C37
 
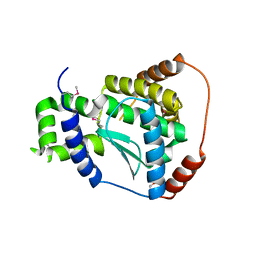 | | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A. Northeast Structural Genomics Consortium target GsR143A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidase, M48 family, ... | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Vorobiev, S.M, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A.
To be Published
|
|
3C5M
 
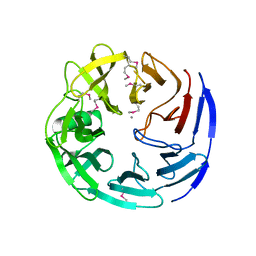 | | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR199 | | Descriptor: | MANGANESE (II) ION, Oligogalacturonate lyase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Owens, L.A, Wang, D, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus.
To be Published
|
|
4V1F
 
 | |
4V9B
 
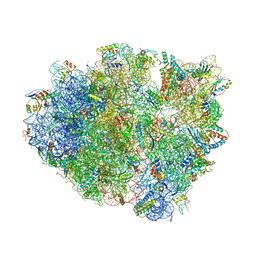 | | Crystal Structure of the 70S ribosome with tigecycline. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2012-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1QV1
 
 | | Atomic resolution structure of obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Liu, Z.J, Vysotski, E.S, Deng, L, Lee, J, Rose, J, Wang, B.C. | | Deposit date: | 2003-08-26 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of obelin: soaking with calcium enhances electron density of the second oxygen atom substituted at the C2-position of coelenterazine.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
4V1G
 
 | |
4UY1
 
 | | Novel pyrazole series of group X Secretory Phospholipase A2 (sPLA2-X) inhibitors | | Descriptor: | 5-(2,5-DIMETHYL-3-THIENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L, Hallberg, K, Bodin, C, Chen, H. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-15 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Pyrazole Series of Group X Secreted Phospholipase A2 Inhibitor (Spla2X) Via Fragment Based Virtual Screening
Bioorg.Med.Chem.Lett., 24, 2014
|
|
