4KSE
 
 | | Crystal structure of a HIV p51 (219-230) deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, HIV p51 subunit | | Authors: | Zheng, X, Mueller, G.A, Derose, E.F, Pedersen, L.C, Gabel, S.A, Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2013-05-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.
Nucleic Acids Res., 42, 2014
|
|
3QVG
 
 | | XRCC1 bound to DNA ligase | | Descriptor: | DNA ligase 3, DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
3PC6
 
 | |
3PC7
 
 | |
3PC8
 
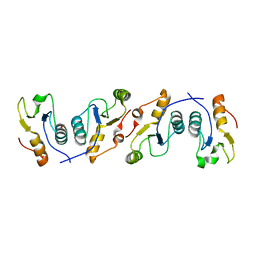 | | X-ray crystal structure of the heterodimeric complex of XRCC1 and DNA ligase III-alpha BRCT domains. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA ligase 3, DNA repair protein XRCC1, ... | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2010-10-21 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
3K4E
 
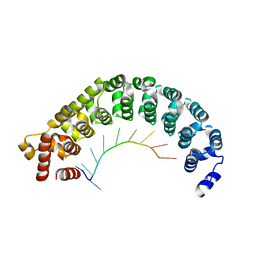 | | Puf3 RNA binding domain bound to Cox17 RNA 3' UTR recognition sequence site A | | Descriptor: | RNA (5'-R(P*CP*UP*UP*GP*UP*AP*UP*AP*UP*A)-3'), mRNA-binding protein PUF3 | | Authors: | Zhu, D, Stumpf, C.R, Krahn, J.M, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K49
 
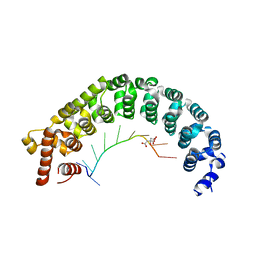 | | Puf3 RNA binding domain bound to Cox17 RNA 3' UTR recognition sequence site B | | Descriptor: | CITRIC ACID, RNA (5'-R(*CP*CP*UP*GP*UP*AP*AP*AP*UP*A)-3'), mRNA-binding protein PUF3 | | Authors: | Zhu, D, Stumpf, C.R, Krahn, J.M, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1ECJ
 
 | |
8V55
 
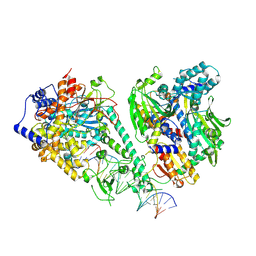 | | Human mitochondrial DNA polymerase gamma bound to a replication fork in an open conformation | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, M.J, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
8V58
 
 | | Complex of murine cathepsin K with bound heparan sulfate 12mer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Pedersen, L.C, Xu, D, Krahn, J.M. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
2P66
 
 | | Human DNA Polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(P*(3DR)P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Prasad, R, Batra, V.K, Yang, X.-P, Krahn, J.M, Pedersen, L.C, Beard, W.A, Wilson, S.H. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the DNA Polymerase beta deoxyribose phosphate lyase mechanism
DNA REPAIR, 4, 2005
|
|
2PFP
 
 | | DNA Polymerase lambda in complex with DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, Downstream Primer, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
2PFQ
 
 | | Manganese promotes catalysis in a DNA polymerase lambda-DNA crystal | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, Downstream Primer, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
2PFN
 
 | | Na in the active site of DNA Polymerase lambda | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA polymerase lambda, Downstream Primer, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
2PFO
 
 | | DNA Polymerase lambda in complex with DNA and dUPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA polymerase lambda, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
1OMZ
 
 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMX
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1ON8
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1ON6
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1YRM
 
 | |
1YZD
 
 | |
1Z7F
 
 | |
1YY0
 
 | | Crystal structure of an RNA duplex containing a 2'-amine substitution and a 2'-amide product produced by in-crystal acylation at a C-A mismatch | | Descriptor: | 5'-R(*GP*CP*AP*GP*AP*(A5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', 5'-R(*GP*CP*AP*GP*AP*(M5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', CALCIUM ION | | Authors: | Gherghe, C.M, Krahn, J.M, Weeks, K.M. | | Deposit date: | 2005-02-23 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures, reactivity and inferred acylation transition States for 2'-amine substituted RNA.
J.Am.Chem.Soc., 127, 2005
|
|
