5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
5GN7
 
 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5GN9
 
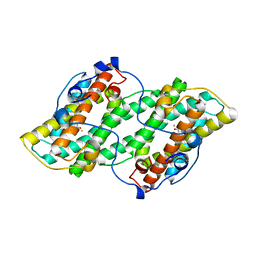 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17b | | Descriptor: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5GN5
 
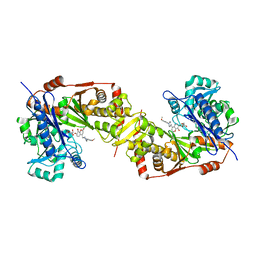 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5C2T
 
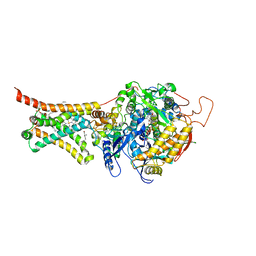 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with rhodoquinone-2 | | Descriptor: | 2-amino-5-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-3-methoxy-6-methylcyclohexa-2,5-diene-1,4-dione, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-06-16 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
1J26
 
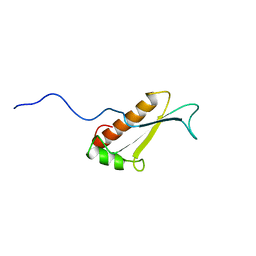 | | Solution structure of a putative peptidyl-tRNA hydrolase domain in a mouse hypothetical protein | | Descriptor: | immature colon carcinoma transcript 1 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality
J.Mol.Biol., 2010
|
|
4YT0
 
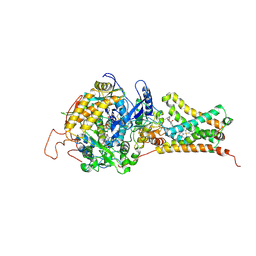 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide. | | Descriptor: | 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YSY
 
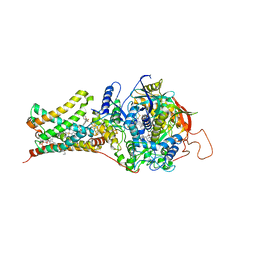 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[(2,4-dichlorophenyl)methyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YSX
 
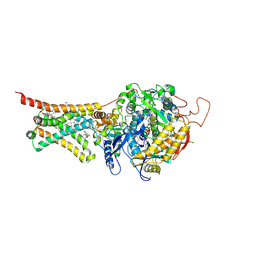 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with the specific inhibitor NN23 | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YTM
 
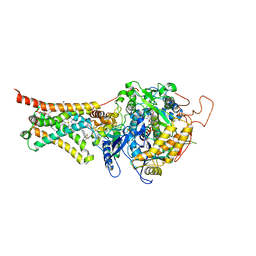 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-biphenyl-3-yl-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YTP
 
 | | CRYSTAL STRUCTURE OF PORCINE HEART MITOCHONDRIAL COMPLEX II BOUND WITH N-[(4-tert-butylphenyl)methyl]-2-(trifluoromethyl)benzamide | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YSZ
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide | | Descriptor: | 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
4YXD
 
 | | CRYSTAL STRUCTURE OF PORCINE HEART MITOCHONDRIAL COMPLEX II BOUND WITH flutolanil | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
5C3J
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with Ubiquinone-1 | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
To Be Published
|
|
4YTN
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[3-(pentafluorophenoxy)phenyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New Insights into the Design of Inhibitors Targeted for Parasitic Anaerobic Energy Metabolism
To Be Published
|
|
2RQC
 
 | | Solution Structure of RNA-binding domain 3 of CUGBP1 in complex with RNA (UG)3 | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*G)-3', CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 37, 2009
|
|
2RRB
 
 | | Refinement of RNA binding domain in human Tra2 beta protein | | Descriptor: | cDNA FLJ40872 fis, clone TUTER2000283, highly similar to Homo sapiens transformer-2-beta (SFRS10) gene | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
1IVZ
 
 | | Solution structure of the SEA domain from murine hypothetical protein homologous to human mucin 16 | | Descriptor: | hypothetical protein 1110008I14RIK | | Authors: | Maeda, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-02 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16)
J.Biol.Chem., 279, 2004
|
|
2YUY
 
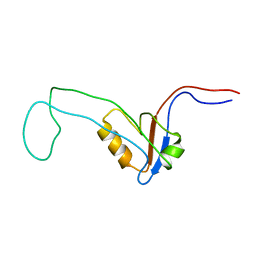 | | Solution Structure of PDZ domain of Rho GTPase Activating Protein 21 | | Descriptor: | Rho GTPase activating protein 21 | | Authors: | Niraula, T.N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of PDZ domain of Rho GTPase Activating Protein 21
To be Published
|
|
2YRK
 
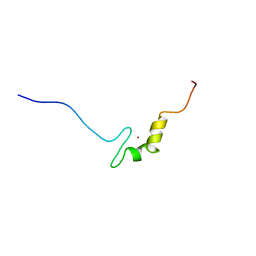 | | Solution structure of the zf-C2H2 domain in zinc finger homeodomain 4 | | Descriptor: | ZINC ION, Zinc finger homeobox protein 4 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-C2H2 domain in zinc finger homeodomain 4
To be Published
|
|
2YU3
 
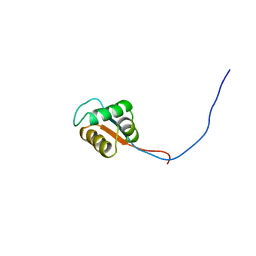 | | Solution structure of the domain swapped WingedHelix in DNA-directed RNA polymerase III 39 kDa polypeptide | | Descriptor: | DNA-directed RNA polymerase III 39 kDa polypeptide F variant | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the domain swapped WingedHelix in DNA-directed RNA polymerase III 39 kDa polypeptide
To be Published
|
|
2YQL
 
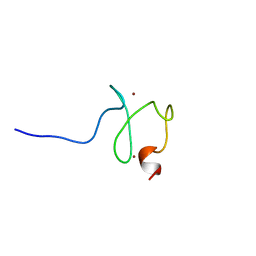 | | Solution structure of the PHD domain in PHD finger protein 21A | | Descriptor: | PHD finger protein 21A, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain in PHD finger protein 21A
To be Published
|
|
2YT9
 
 | | Solution structure of C2H2 type Zinc finger domain 345 in Zinc finger protein 278 | | Descriptor: | ZINC ION, zinc finger-containing protein 1 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 345 in Zinc finger protein 278
To be Published
|
|
2YQR
 
 | | Solution structure of the KH domain in KIAA0907 protein | | Descriptor: | KIAA0907 protein | | Authors: | Kadirvel, S, He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KH domain in KIAA0907 protein
To be Published
|
|
2YT6
 
 | | Solution structure of the SH3_1 domain of Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 | | Descriptor: | Adult male urinary bladder cDNA, RIKEN full-length enriched library, clone:9530076O17 product:Yamaguchi sarcoma viral (v-yes) oncogene homolog | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3_1 domain of Yamaguchi sarcoma viral (v-yes) oncogene homolog 1
To be Published
|
|
