6N4Y
 
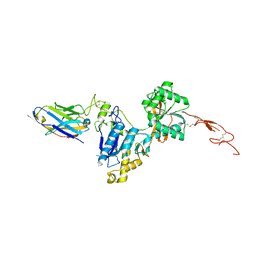 | | Metabotropic Glutamate Receptor 5 Extracellular Domain with Nb43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Mathiesen, J.M, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.262 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6OYA
 
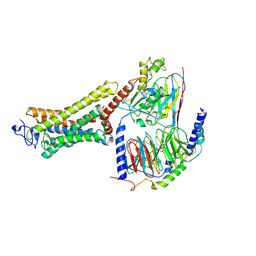 | | Structure of the Rhodopsin-Transducin-Nanobody Complex | | Descriptor: | Camelid antibody VHH fragment, Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
6N52
 
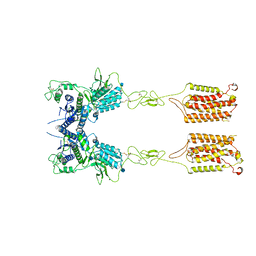 | | Metabotropic Glutamate Receptor 5 Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N51
 
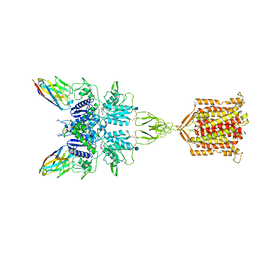 | | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
2B7D
 
 | | Factor VIIa Inhibitors: Chemical Optimization, Preclinical Pharmacokinetics, Pharmacodynamics, and Efficacy in a Baboon Thrombosis Model | | Descriptor: | (2R)-2-[5-(5-CARBAMIMIDOYL-1H-BENZOIMIDAZOL-2-YL)-6,2'-DIHYDROXY-5'-UREIDOMETHYL-BIPHENYL-3-YL]-SUCCINIC ACID, Coagulation factor VII, Tissue factor | | Authors: | Young, W.B, Mordenti, J, Torkelson, S, Shrader, W.D, Kolesnikov, A, Rai, R, Liu, L, Hu, H, Leahy, E.M, Green, M.J, Sprengeler, P.A, Katz, B.A, Yu, C, Janc, J.W, Elrod, K.C, Marzec, U.M, Hanson, S.R. | | Deposit date: | 2005-10-04 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Factor VIIa inhibitors: Chemical optimization, preclinical pharmacokinetics, pharmacodynamics, and efficacy in an arterial baboon thrombosis model.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3EOP
 
 | | Crystal Structure of the DUF55 domain of human thymocyte nuclear protein 1 | | Descriptor: | SULFATE ION, Thymocyte nuclear protein 1 | | Authors: | Yu, F, Song, A, Xu, C, Sun, L, Li, L, Tang, L, Hu, H, He, J. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determining the DUF55-domain structure of human thymocyte nuclear protein 1 from crystals partially twinned by tetartohedry
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2FLR
 
 | | Novel 5-Azaindole Factor VIIa Inhibitors | | Descriptor: | Coagulation factor VII, Tissue factor, [2'-HYDROXY-3'-(1H-PYRROLO[3,2-C]PYRIDIN-2-YL)-BIPHENYL-3-YLMETHYL]-UREA | | Authors: | Riggs, J.R, Hu, H, Kolesnikov, A, Tong, Z, Leahy, E.M, Wesson, K.E, Shrader, W.D, Vijaykumar, D, Wahl, T.A, Sprengeler, P.A, Green, M.J, Yu, C, Katz, B.A, Young, W.B. | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel 5-azaindole factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5VAI
 
 | | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein | | Descriptor: | Glucagon-like peptide 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y, Sun, B, Feng, D, Hu, H, Chu, M, Qu, Q, Tarrasch, J.T, Li, S, Kobilka, T.S, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2017-03-27 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Nature, 546, 2017
|
|
8IHO
 
 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
2LJ1
 
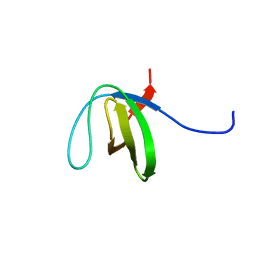 | | The third SH3 domain of R85FL with ataxin-7 PRR | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
2LBC
 
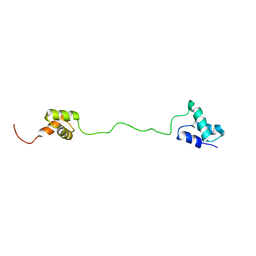 | | solution structure of tandem UBA of USP13 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 13 | | Authors: | Zhang, Y, Zhou, C, Zhou, Z, Song, A, Hu, H. | | Deposit date: | 2011-03-29 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain Analysis Reveals That a Deubiquitinating Enzyme USP13 Performs Non-Activating Catalysis for Lys63-Linked Polyubiquitin.
Plos One, 6, 2011
|
|
2LJ0
 
 | | The third SH3 domain of R85FL | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
2L5F
 
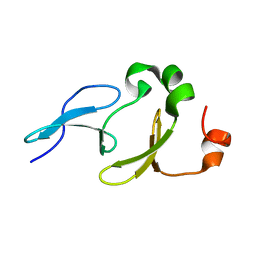 | | Solution structure of the tandem WW domains from HYPA/FBP11 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interaction with polyglutamine expanded huntingtin alters cellular distribution and RNA processing of huntingtin yeast two-hybrid protein A (HYPA)
To be Published
|
|
2L80
 
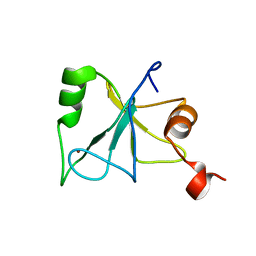 | | Solution Structure of the Zinc Finger Domain of USP13 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 13, ZINC ION | | Authors: | Zhang, Y, Zhou, C, Zhou, Z, Song, A, Hu, H. | | Deposit date: | 2010-12-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biochemical Characterization of the Ubiquitin Receptors in USP13 Reveals Different Catalytic Activation of Deubiquitination from Its Analogue USP5
To be Published
|
|
2L5V
 
 | |
2KSN
 
 | | Solution Structure of the N-terminal Domain of DC-UbP/UBTD2 | | Descriptor: | Ubiquitin domain-containing protein 2 | | Authors: | Song, A, Zhou, C, Guan, X, Sze, K, Hu, H. | | Deposit date: | 2010-01-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of DC-UbP/UBTD2 and its interaction with ubiquitin
Protein Sci., 19, 2010
|
|
8HIL
 
 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | Descriptor: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | Authors: | Du, X, Xie, G, Hu, H, Du, J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8GYD
 
 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
6JPD
 
 | | Mouse receptor-interacting protein kinase 3 (RIP3) amyloid structure by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Hu, H, Zhang, J, Dong, X.Q, Wang, J, Schwieters, C, Wang, H.Y, Lu, J.X. | | Deposit date: | 2019-03-26 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Nat Commun, 12, 2021
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5YYS
 
 | | Cryo-EM structure of L-fucokinase, GDP-fucose pyrophosphorylase (FKP)in Bacteroides fragilis | | Descriptor: | L-fucokinase, L-fucose-1-P guanylyltransferase | | Authors: | Wang, J, Hu, H, Liu, Y, Zhou, Q, Wu, P, Yan, N, Wang, H.W, Wu, J.W, Sun, L. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of L-fucokinase/GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis.
Protein Cell, 10, 2019
|
|
7EQ4
 
 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
6V4B
 
 | | DeCLIC N-terminal Domain 34-202 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Neur_chan_LBD domain-containing protein, ... | | Authors: | Delarue, M, Hu, H.D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for allosteric transitions of a multidomain pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V4A
 
 | |
