7OXQ
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA in complex with fragment 048 at the transient P-pocket. | | Descriptor: | 2-(4-bromanylpyrazol-1-yl)-~{N}-cyclopropyl-~{N}-methyl-ethanamide, CADMIUM ION, DNA (28-MER), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
7OZ2
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA showing a transient P-pocket | | Descriptor: | CADMIUM ION, DNA (28-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
7OZW
 
 | |
7OZ5
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA in complex with fragment 166 at the transient P-pocket. | | Descriptor: | (1~{R},2~{R})-2-phenyl-~{N}-(1,3-thiazol-2-yl)cyclopropane-1-carboxamide, CADMIUM ION, DNA (28-MER), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
5JUN
 
 | | PB2 bound to an azaindole inhibitor | | Descriptor: | (3~{R})-3-[[5-fluoranyl-2-(5-fluoranyl-1~{H}-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]-3-(1-methylcyclobutyl)propanoic acid, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel, Orally Bioavailable beta-Amino Acid Azaindole Inhibitors of Influenza PB2.
ACS Med Chem Lett, 8, 2017
|
|
7NAI
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with 3AD | | Descriptor: | Sterile alpha and TIR motif-containing protein 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAJ
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ara-2'F-ADPR | | Descriptor: | 1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-D-arabinitol, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Shi, Y, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAG
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with 1AD | | Descriptor: | Sterile alpha and TIR motif-containing protein 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Bosanac, T, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAH
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with 2AD | | Descriptor: | Sterile alpha and TIR motif-containing protein 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(8-oxidanylidene-7~{H}-2,7-naphthyridin-2-yl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
5JUR
 
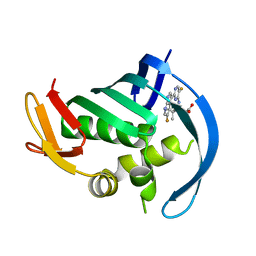 | | PB2 bound to an azaindole inhibitor | | Descriptor: | (3~{R})-3-[[5-fluoranyl-2-(5-fluoranyl-1~{H}-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]-4,4-dimethyl-pentanoic acid, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of Novel, Orally Bioavailable beta-Amino Acid Azaindole Inhibitors of Influenza PB2.
ACS Med Chem Lett, 8, 2017
|
|
8BTS
 
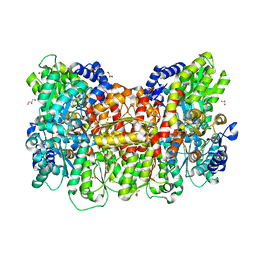 | |
8D0I
 
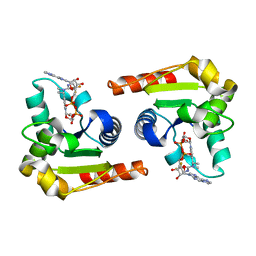 | | Human SARM1 bound to an NB-3 eADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0C
 
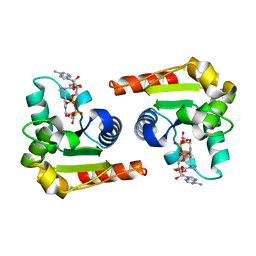 | | Human SARM1 TIR domain bound to NB-3-ADPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0G
 
 | | Human SARM1 TIR domain bound to NB-3-ADPRP | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0D
 
 | | Human SARM1 TIR domain bound to an NB-7-ADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R})-5-[4-[3-[3-(4-chlorophenyl)propanoylamino]-4-methyl-1~{H}-pyrazol-5-yl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0H
 
 | | Human SARM1 TIR domain bound to NB-3-GDPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0F
 
 | | Human SARM1 TIR domain bound to NB-2-ADPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]methyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0E
 
 | | Human SARM1 TIR domain bound to NB-7 | | Descriptor: | 3-(4-chlorophenyl)-N-[4-methyl-3-(pyridin-4-yl)-1H-pyrazol-5-yl]propanamide, NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0J
 
 | | Apo Human SARM1 TIR domain | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8K9R
 
 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9Q
 
 | | Cryo-EM structure of the GPI inositol-deacylase (PGAP1/Bst1) from Chaetomium thermophilum | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, CHOLESTEROL HEMISUCCINATE, GPI inositol-deacylase,fused thermostable green fluorescent protein | | Authors: | Hong, J, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9T
 
 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
4YD0
 
 | | Influenza polymerase basic protein 2 (PB2) bound to an azaindole-tetrazole inhibitor | | Descriptor: | 2-(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-fluoro-N-[(1R,2S,3S,4R)-3-(1H-tetrazol-5-yl)bicyclo[2.2.2]oct-2-yl]pyrimidin-4-amine, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Isosteric replacements of the carboxylic acid of drug candidate VX-787: Effect of charge on antiviral potency and kinase activity of azaindole-based influenza PB2 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
