7CDE
 
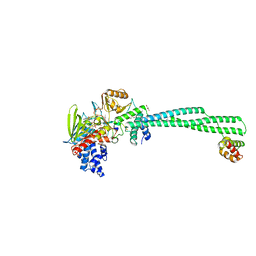 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRKR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
5YJP
 
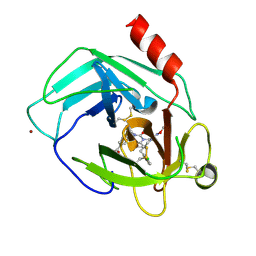 | | Human chymase in complex with 3-(ethoxyimino)-7-oxo-1,4-diazepane derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-((R)-1-((R,Z)-6-(5-chloro-2-methoxybenzyl)-3-(ethoxyimino)-7-oxo-1,4-diazepane-1-carboxamido)propyl)benzoic acid, ZINC ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5YJM
 
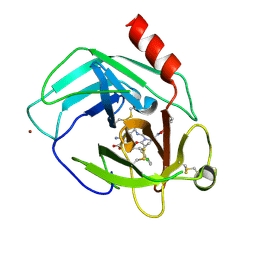 | | Human chymase in complex with 7-oxo-3-(phenoxyimino)-1,4-diazepane derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-4-((R)-1-((R,Z)-6-(5-chloro-2-methoxybenzyl)-7-oxo-3-(phenoxyimino)-1,4-diazepane-1-carboxamido)propyl)benzoic acid, ZINC ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7WYT
 
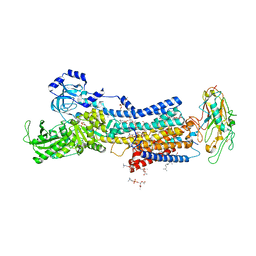 | | Crystal structures of Na+,K+-ATPase in complex with ouabain | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYS
 
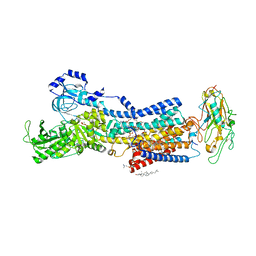 | | Crystal structures of Na+,K+-ATPase in complex with istaroxime | | Descriptor: | (3E,5S,8R,9S,10R,13S,14S)-3-(2-azanylethoxyimino)-10,13-dimethyl-1,2,4,5,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthrene-6,17-dione, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DDF
 
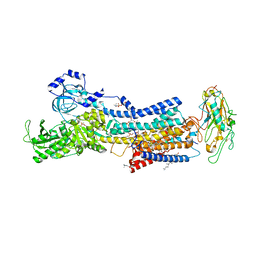 | | Crystal structures of Na+,K+-ATPase in complex with beryllium fluoride | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.62 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DDL
 
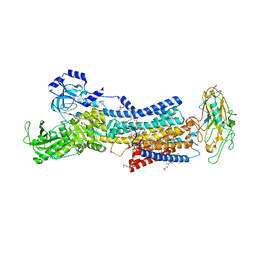 | | Crystal structures of Na+,K+-ATPase in complex with bufalin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DDH
 
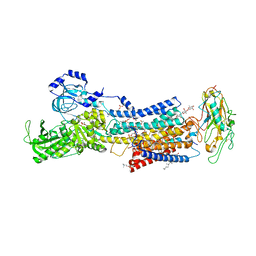 | | Crystal structures of Na+,K+-ATPase in complex with digoxin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DDI
 
 | | Crystal structures of Na+,K+-ATPase in complex with digitoxin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DDK
 
 | | Crystal structures of Na+,K+-ATPase in complex with rostafuroxin | | Descriptor: | (3S,5R,8R,9S,10S,13S,14S,17S)-17-(furan-3-yl)-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,15,16-dodecahydro-1H-cyclopenta[a]phenanthrene-3,14,17-triol, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3CU0
 
 | | human beta 1,3-glucuronyltransferase I (GlcAT-I) in complex with UDP and GAL-GAL(6-SO4)-XYL(2-PO4)-O-SER | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 3, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Tone, Y, Pedersen, L.C, Yamamoto, T, Kitagawa, H, Nishihara-Shimizu, J, Tamura, J, Negishi, M, Sugahara, K. | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-o-phosphorylation of xylose and 6-o-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis.
J.Biol.Chem., 283, 2008
|
|
1BXU
 
 | | OXIDIZED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | Deposit date: | 1998-10-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
1D4F
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT RAT-LIVER D244E MUTANT S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Komoto, J, Huang, Y, Takusagawa, F, Gomi, T, Ogawa, H, Takata, Y, Fujioka, M. | | Deposit date: | 2000-06-22 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Effects of site-directed mutagenesis on structure and function of recombinant rat liver S-adenosylhomocysteine hydrolase. Crystal structure of D244E mutant enzyme.
J.Biol.Chem., 275, 2000
|
|
1QSI
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), ... | | Authors: | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | Deposit date: | 1999-06-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
1QSH
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | Deposit date: | 1999-06-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
1C7Y
 
 | | E.COLI RUVA-HOLLIDAY JUNCTION COMPLEX | | Descriptor: | DNA (5'-D(P*DAP*DAP*DGP*DTP*DTP*DGP*DGP*DGP*DAP*DTP*DTP*DGP*DT)-3'), DNA (5'-D(P*DCP*DAP*DAP*DTP*DCP*DCP*DCP*DAP*DAP*DCP*DTP*DT)-3'), DNA (5'-D(P*DCP*DGP*DAP*DAP*DTP*DGP*DTP*DGP*DTP*DGP*DTP*DCP*DT)-3'), ... | | Authors: | Ariyoshi, M, Nishino, T, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1D8L
 
 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
1IPB
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GPPPA | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
1IPC
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
1IXS
 
 | | Structure of RuvB complexed with RuvA domain III | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1IWA
 
 | | RUBISCO FROM GALDIERIA PARTITA | | Descriptor: | SULFATE ION, ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | | Authors: | Okano, Y, Mizohata, E, Xie, Y, Matsumura, H, Sugawara, H, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2002-04-30 | | Release date: | 2003-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Galdieria Rubisco Complexed with one sulfate ion per active site
FEBS LETT., 527, 2002
|
|
1IXR
 
 | | RuvA-RuvB complex | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1BXV
 
 | | REDUCED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | Deposit date: | 1998-10-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
2RKB
 
 | | Serine dehydratase like-1 from human cancer cells | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine dehydratase-like | | Authors: | Yamada, T, Komoto, J, Kasuya, T, Mori, H, Ogawa, H, Takusagawa, F. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A catalytic mechanism that explains a low catalytic activity of serine dehydratase like-1 from human cancer cells: Crystal structure and site-directed mutagenesis studies.
Biochim.Biophys.Acta, 1780, 2008
|
|
1XWF
 
 | | K185N mutated S-adenosylhomocysteine hydrolase | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, T, Takata, Y, Komoto, J, Gomi, T, Ogawa, H, Fujioka, M, Takusagawa, F. | | Deposit date: | 2004-11-01 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism of S-adenosylhomocysteine hydrolase: Roles of His 54, Asp130, Glu155, Lys185, and Aspl89.
Int.J.Biochem.Cell Biol., 37, 2005
|
|
