7FHO
 
 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 50 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHL
 
 | | Structure of AtTPC1 with 50 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHK
 
 | | Structure of AtTPC1 with 1 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8T21
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T22
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T25
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation. | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T20
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T23
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8TAZ
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Liang, B. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8DTS
 
 | |
2G88
 
 | | MSRECA-dATP COMPLEX | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Krishna, R, Manjunath, G.P, Kumar, P, Surolia, A, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-03-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic identification of an ordered C-terminal domain and a second nucleotide-binding site in RecA: new insights into allostery.
Nucleic Acids Res., 34, 2006
|
|
6URI
 
 | |
2F7T
 
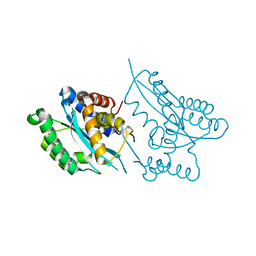 | | Crystal structure of the catalytic domain of Mos1 mariner transposase | | Descriptor: | MAGNESIUM ION, Mos1 transposase | | Authors: | Richardson, J.M, Dawson, A, Taylor, P, Finnegan, D.J, Walkinshaw, M.D. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Mos1 transposition: insights from structural analysis
Embo J., 25, 2006
|
|
7S6I
 
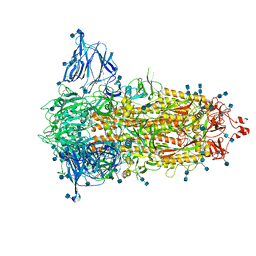 | | SARS-CoV-2-6P-Mut2 S protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7S6J
 
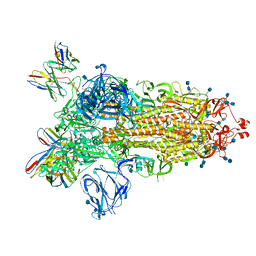 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7S6L
 
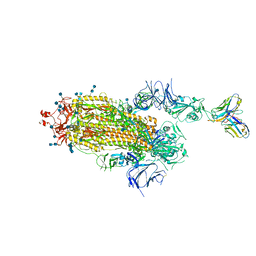 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (conformation 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7S6K
 
 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SBU
 
 | |
7RRW
 
 | | Monomeric CRM197 expressed in E. coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diphtheria toxin | | Authors: | Gallagher, D.T, Lees, A. | | Deposit date: | 2021-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric crystal structure of the vaccine carrier protein CRM 197 and implications for vaccine development.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
2OFO
 
 | | MSrecA-native | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2007-01-04 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2OEP
 
 | | MSrecA-ADP-complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2ODW
 
 | | MSrecA-ATP-GAMA-S complex | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2ODN
 
 | | MSRECA-dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-24 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2OE2
 
 | | MSrecA-native-low humidity 95% | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
6Z8W
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3G), which contains 1-beta-D-glucopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
